[English] 日本語
 Yorodumi
Yorodumi- EMDB-34919: CryoEM structure of HpaCas9-sgRNA-dsDNA in the presence of AcrIIC4 -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM structure of HpaCas9-sgRNA-dsDNA in the presence of AcrIIC4 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Cas9 / cleavage inhibition / ANTIMICROBIAL PROTEIN / HYDROLASE-RNA-ANTIMICROBIAL PROTEIN complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationmaintenance of CRISPR repeat elements / endonuclease activity / defense response to virus / Hydrolases; Acting on ester bonds / DNA binding / RNA binding / metal ion binding Similarity search - Function | |||||||||
| Biological species |  Haemophilus parainfluenzae (bacteria) / synthetic construct (others) Haemophilus parainfluenzae (bacteria) / synthetic construct (others) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.1 Å | |||||||||
 Authors Authors | Sun W / Cheng Z / Wang J / Yang X / Wang Y | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation | Journal: mBio / Year: 2018 Title: Potent Cas9 Inhibition in Bacterial and Human Cells by AcrIIC4 and AcrIIC5 Anti-CRISPR Proteins. Authors: Jooyoung Lee / Aamir Mir / Alireza Edraki / Bianca Garcia / Nadia Amrani / Hannah E Lou / Ildar Gainetdinov / April Pawluk / Raed Ibraheim / Xin D Gao / Pengpeng Liu / Alan R Davidson / ...Authors: Jooyoung Lee / Aamir Mir / Alireza Edraki / Bianca Garcia / Nadia Amrani / Hannah E Lou / Ildar Gainetdinov / April Pawluk / Raed Ibraheim / Xin D Gao / Pengpeng Liu / Alan R Davidson / Karen L Maxwell / Erik J Sontheimer /   Abstract: In their natural settings, CRISPR-Cas systems play crucial roles in bacterial and archaeal adaptive immunity to protect against phages and other mobile genetic elements, and they are also widely used ...In their natural settings, CRISPR-Cas systems play crucial roles in bacterial and archaeal adaptive immunity to protect against phages and other mobile genetic elements, and they are also widely used as genome engineering technologies. Previously we discovered bacteriophage-encoded Cas9-specific anti-CRISPR (Acr) proteins that serve as countermeasures against host bacterial immunity by inactivating their CRISPR-Cas systems (A. Pawluk, N. Amrani, Y. Zhang, B. Garcia, et al., Cell 167:1829-1838.e9, 2016, https://doi.org/10.1016/j.cell.2016.11.017). We hypothesized that the evolutionary advantages conferred by anti-CRISPRs would drive the widespread occurrence of these proteins in nature (K. L. Maxwell, Mol Cell 68:8-14, 2017, https://doi.org/10.1016/j.molcel.2017.09.002; A. Pawluk, A. R. Davidson, and K. L. Maxwell, Nat Rev Microbiol 16:12-17, 2018, https://doi.org/10.1038/nrmicro.2017.120; E. J. Sontheimer and A. R. Davidson, Curr Opin Microbiol 37:120-127, 2017, https://doi.org/10.1016/j.mib.2017.06.003). We have identified new anti-CRISPRs using the same bioinformatic approach that successfully identified previous Acr proteins (A. Pawluk, N. Amrani, Y. Zhang, B. Garcia, et al., Cell 167:1829-1838.e9, 2016, https://doi.org/10.1016/j.cell.2016.11.017) against Cas9 (NmeCas9). In this work, we report two novel anti-CRISPR families in strains of and , both of which harbor type II-C CRISPR-Cas systems (A. Mir, A. Edraki, J. Lee, and E. J. Sontheimer, ACS Chem Biol 13:357-365, 2018, https://doi.org/10.1021/acschembio.7b00855). We characterize the type II-C Cas9 orthologs from and , show that the newly identified Acrs are able to inhibit these systems, and define important features of their inhibitory mechanisms. The Acr is the most potent NmeCas9 inhibitor identified to date. Although inhibition of NmeCas9 by anti-CRISPRs from and reveals cross-species inhibitory activity, more distantly related type II-C Cas9s are not inhibited by these proteins. The specificities of anti-CRISPRs and divergent Cas9s appear to reflect coevolution of their strategies to combat or evade each other. Finally, we validate these new anti-CRISPR proteins as potent off-switches for Cas9 genome engineering applications. As one of their countermeasures against CRISPR-Cas immunity, bacteriophages have evolved natural inhibitors known as anti-CRISPR (Acr) proteins. Despite the existence of such examples for type II CRISPR-Cas systems, we currently know relatively little about the breadth of Cas9 inhibitors, and most of their direct Cas9 targets are uncharacterized. In this work we identify two new type II-C anti-CRISPRs and their cognate Cas9 orthologs, validate their functionality and in bacteria, define their inhibitory spectrum against a panel of Cas9 orthologs, demonstrate that they act before Cas9 DNA binding, and document their utility as off-switches for Cas9-based tools in mammalian applications. The discovery of diverse anti-CRISPRs, the mechanistic analysis of their cognate Cas9s, and the definition of Acr inhibitory mechanisms afford deeper insight into the interplay between Cas9 orthologs and their inhibitors and provide greater scope for exploiting Acrs for CRISPR-based genome engineering. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_34919.map.gz emd_34919.map.gz | 22.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-34919-v30.xml emd-34919-v30.xml emd-34919.xml emd-34919.xml | 22.6 KB 22.6 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_34919.png emd_34919.png | 138.6 KB | ||
| Filedesc metadata |  emd-34919.cif.gz emd-34919.cif.gz | 7.3 KB | ||
| Others |  emd_34919_half_map_1.map.gz emd_34919_half_map_1.map.gz emd_34919_half_map_2.map.gz emd_34919_half_map_2.map.gz | 18.1 MB 18.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-34919 http://ftp.pdbj.org/pub/emdb/structures/EMD-34919 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34919 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34919 | HTTPS FTP |
-Validation report
| Summary document |  emd_34919_validation.pdf.gz emd_34919_validation.pdf.gz | 914.3 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_34919_full_validation.pdf.gz emd_34919_full_validation.pdf.gz | 913.9 KB | Display | |
| Data in XML |  emd_34919_validation.xml.gz emd_34919_validation.xml.gz | 10.1 KB | Display | |
| Data in CIF |  emd_34919_validation.cif.gz emd_34919_validation.cif.gz | 11.8 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34919 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34919 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34919 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34919 | HTTPS FTP |
-Related structure data
| Related structure data |  8hnvMC  8hnsC  8hntC  8hnwC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_34919.map.gz / Format: CCP4 / Size: 23.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_34919.map.gz / Format: CCP4 / Size: 23.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.04 Å | ||||||||||||||||||||||||||||||||||||
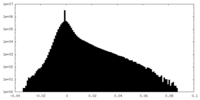
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_34919_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
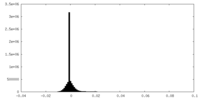
| Density Histograms |
-Half map: #1
| File | emd_34919_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
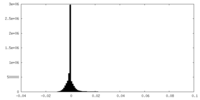
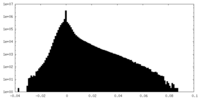
| Density Histograms |
- Sample components
Sample components
-Entire : Complex of HpaCas9-sgRNA-DNA in the presence of AcrIIC4
| Entire | Name: Complex of HpaCas9-sgRNA-DNA in the presence of AcrIIC4 |
|---|---|
| Components |
|
-Supramolecule #1: Complex of HpaCas9-sgRNA-DNA in the presence of AcrIIC4
| Supramolecule | Name: Complex of HpaCas9-sgRNA-DNA in the presence of AcrIIC4 type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#5 |
|---|---|
| Source (natural) | Organism:  Haemophilus parainfluenzae (bacteria) Haemophilus parainfluenzae (bacteria) |
| Molecular weight | Theoretical: 194 KDa |
-Supramolecule #2: HpaCas9/AcrIIC4
| Supramolecule | Name: HpaCas9/AcrIIC4 / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1, #5 |
|---|
-Supramolecule #3: RNA/DNA
| Supramolecule | Name: RNA/DNA / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2-#4 |
|---|
-Macromolecule #1: CRISPR-associated endonuclease Cas9
| Macromolecule | Name: CRISPR-associated endonuclease Cas9 / type: protein_or_peptide / ID: 1 Details: Two mutations D13A and H581A were introduced to inactivate the catalytic sites of HpaCas9. The first residue 'Ser' of the sample sequence is the one expressed from the vector left after tag cleavage. Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Haemophilus parainfluenzae (bacteria) Haemophilus parainfluenzae (bacteria) |
| Molecular weight | Theoretical: 121.605695 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: SMENKNLNYI LGLALGIASV GWAVVEIDEK ENPLRLIDVG VRTFERAEVP KTGESLALSR RLARSARRLT QRRVARLKKA KRLLKSENI LLSTDERLPH QVWQLRVEGL DHKLERQEWA AVLLHLIKHR GYLSQRKNES KSENKELGAL LSGVDNNHKL L QQATYRSP ...String: SMENKNLNYI LGLALGIASV GWAVVEIDEK ENPLRLIDVG VRTFERAEVP KTGESLALSR RLARSARRLT QRRVARLKKA KRLLKSENI LLSTDERLPH QVWQLRVEGL DHKLERQEWA AVLLHLIKHR GYLSQRKNES KSENKELGAL LSGVDNNHKL L QQATYRSP AELAVKKFEV EEGHIRNQQG AYTHTFSRLD LLAEMELLFS RQQHFGNPFA SEKLLENLTA LLMWQKPALS GE AILKMLG KCTFEDEYKA AKNTYSAERF VWITKLNNLR IQENGLERAL NDNERLALME QPYDKNRLFY SQVRSILKLS DEA IFKGLR YSGEDKKAIE TKAVLMEMKA YHQIRKVLEG NNLKAEWAEL KANPTLLDEI GTAFSLYKTD EDISAYLAGK LSQP VLNAL LENLSFDKFI QLSLKALYKL LPLMQQGLRY DEACREIYGD HYGKKTEENH HFLPQIPADE IRNPVVLRTL TQARK VING VVRLYGSPAR IHIETGREVG KSYKDRRELE KRQEENRKQR ENAIKEFKEY FPHFAGEPKA KDILKMRLYK QQNAKC LYS GKPIELHRLL EKGYVEVDAA LPFSRTWDDS FNNKVLVLAN ENQNKGNLTP FEWLDGKHNS ERWRAFKALV ETSAFPY AK KQRILSQKLD EKGFIERNLN DTRYVARFLC NFIADNMHLT GEGKRKVFAS NGQITALLRS RWGLAKSRED NDRHHALD A VVVACSTVAM QQKITRFVRF EAGDVFTGER IDRETGEIIP LHFPTPWQFF KQEVEIRIFS DNPKLELENR LPDRPQANH EFVQPLFVSR MPTRKMTGQG HMETVKSAKR LNEGISVIKM PLTKLKLKDL ELMVNREREK DLYDTLKARL EAFNDDPAKA FAEPFIKKG GAIVKSVRVE QIQKSGVLVR EGNGVADNAS MVRVDVFTKG GKYFLVPIYT WQVAKGILPN KAATQYKDEE D WEVMDNSA TFKFSLHPND LVKLVTKKKT ILGYFNGLNR ATGNIDIKEH DLDKSKGKQG IFEGVGIKLA LSFEKYQVDE LG KNIRLCK PSKRQPVR UniProtKB: CRISPR-associated endonuclease Cas9 |
-Macromolecule #5: anti-CRISPR protein AcrIIC4
| Macromolecule | Name: anti-CRISPR protein AcrIIC4 / type: protein_or_peptide / ID: 5 Details: WP_049372635.1;The first residue 'Ser' of the sample sequence is the one expressed from the vector left after tag cleavage. Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Haemophilus parainfluenzae (bacteria) Haemophilus parainfluenzae (bacteria) |
| Molecular weight | Theoretical: 10.072395 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: SMKITSSNFA TIATSENFAK LSVLPKNHRE PIKGLFKSAV EQFSSARDFF KNENYSKELA EKFNKEAVNE AVEKLQKAID LAEKQGIQF |
-Macromolecule #2: sgRNA
| Macromolecule | Name: sgRNA / type: rna / ID: 2 Details: RNA is originally derived from Haemophilus parainfluenzae and modified by author. Number of copies: 1 |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 40.860125 KDa |
| Sequence | String: GGUCACUCUA ACAUUUAAUC ACACGUUGUA GCUCCCUUUU UCGAAAGAAA AACGUUGUUA CAAUAAGAGA AAAGAUUUCU CGCAAAGCU CUGUCCCUUG AAAUGUAAGU UUCAAGGGAC AUCUUUUUC |
-Macromolecule #3: target strand
| Macromolecule | Name: target strand / type: dna / ID: 3 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism:  Haemophilus parainfluenzae (bacteria) Haemophilus parainfluenzae (bacteria) |
| Molecular weight | Theoretical: 10.865029 KDa |
| Sequence | String: (DT)(DG)(DA)(DA)(DA)(DT)(DC)(DA)(DT)(DA) (DT)(DG)(DT)(DG)(DT)(DG)(DA)(DT)(DT)(DA) (DA)(DA)(DT)(DG)(DT)(DT)(DA)(DG)(DA) (DG)(DT)(DG)(DA)(DC)(DC) |
-Macromolecule #4: non-target strand
| Macromolecule | Name: non-target strand / type: dna / ID: 4 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism:  Haemophilus parainfluenzae (bacteria) Haemophilus parainfluenzae (bacteria) |
| Molecular weight | Theoretical: 10.766954 KDa |
| Sequence | String: (DG)(DG)(DT)(DC)(DA)(DC)(DT)(DC)(DT)(DA) (DA)(DC)(DA)(DT)(DT)(DT)(DA)(DA)(DT)(DG) (DT)(DG)(DT)(DG)(DA)(DT)(DA)(DT)(DG) (DA)(DT)(DT)(DT)(DC)(DA) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 Component:
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.6 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: PDB ENTRY PDB model - PDB ID: |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.1 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 212049 |
| Initial angle assignment | Type: PROJECTION MATCHING |
| Final angle assignment | Type: ANGULAR RECONSTITUTION |
-Atomic model buiding 1
| Refinement | Protocol: RIGID BODY FIT |
|---|---|
| Output model |  PDB-8hnv: |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)