+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Bry-LHCII homotrimer of Bryopsis corticulans | ||||||||||||||||||||||||||||||
 Map data Map data | |||||||||||||||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||||||||||||||
 Keywords Keywords | light-harvesting complexes / siphonaxanthin / siphonein / PHOTOSYNTHESIS | ||||||||||||||||||||||||||||||
| Biological species |  Bryopsis corticulans (plant) Bryopsis corticulans (plant) | ||||||||||||||||||||||||||||||
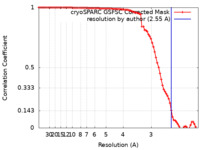
| Method | single particle reconstruction / cryo EM / Resolution: 2.55 Å | ||||||||||||||||||||||||||||||
 Authors Authors | Li ZH / Shen JR / Wang WD | ||||||||||||||||||||||||||||||
| Funding support |  China, 9 items China, 9 items
| ||||||||||||||||||||||||||||||
 Citation Citation |  Journal: Structure / Year: 2023 Journal: Structure / Year: 2023Title: Structural and functional properties of different types of siphonous LHCII trimers from an intertidal green alga Bryopsis corticulans. Authors: Zhenhua Li / Cuicui Zhou / Songhao Zhao / Jinyang Zhang / Xueyang Liu / Min Sang / Xiaochun Qin / Yanyan Yang / Guangye Han / Tingyun Kuang / Jian-Ren Shen / Wenda Wang /   Abstract: Light-harvesting complexes of photosystem II (LHCIIs) in green algae and plants are vital antenna apparatus for light harvesting, energy transfer, and photoprotection. Here we determined the ...Light-harvesting complexes of photosystem II (LHCIIs) in green algae and plants are vital antenna apparatus for light harvesting, energy transfer, and photoprotection. Here we determined the structure of a siphonous-type LHCII trimer from the intertidal green alga Bryopsis corticulans by X-ray crystallography and cryo-electron microscopy (cryo-EM), and analyzed its functional properties by spectral analysis. The Bryopsis LHCII (Bry-LHCII) structures in both homotrimeric and heterotrimeric form show that green light-absorbing siphonaxanthin and siphonein occupied the sites of lutein and violaxanthin in plant LHCII, and two extra chlorophylls (Chls) b replaced Chls a. Binding of these pigments expands the blue-green light absorption of B. corticulans in the tidal zone. We observed differences between the Bry-LHCII homotrimer crystal and cryo-EM structures, and also between Bry-LHCII homotrimer and heterotrimer cryo-EM structures. These conformational changes may reflect the flexibility of Bry-LHCII, which may be required to adapt to light fluctuations from tidal rhythms. | ||||||||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_34883.map.gz emd_34883.map.gz | 32.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-34883-v30.xml emd-34883-v30.xml emd-34883.xml emd-34883.xml | 19.8 KB 19.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_34883_fsc.xml emd_34883_fsc.xml | 8.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_34883.png emd_34883.png | 125.8 KB | ||
| Filedesc metadata |  emd-34883.cif.gz emd-34883.cif.gz | 6.1 KB | ||
| Others |  emd_34883_half_map_1.map.gz emd_34883_half_map_1.map.gz emd_34883_half_map_2.map.gz emd_34883_half_map_2.map.gz | 59.5 MB 59.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-34883 http://ftp.pdbj.org/pub/emdb/structures/EMD-34883 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34883 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34883 | HTTPS FTP |
-Related structure data
| Related structure data |  8hlvMC  8hpdC  8hq8C M: atomic model generated by this map C: citing same article ( |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_34883.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_34883.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.04 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_34883_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
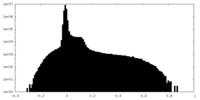
| Projections & Slices |
| ||||||||||||
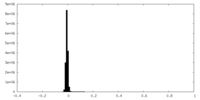
| Density Histograms |
-Half map: #1
| File | emd_34883_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
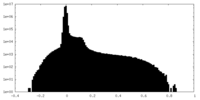
| Projections & Slices |
| ||||||||||||
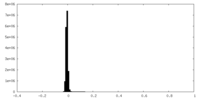
| Density Histograms |
- Sample components
Sample components
-Entire : Bryopsis corticulans chlorophyll a/b-type light-harvesting comple...
| Entire | Name: Bryopsis corticulans chlorophyll a/b-type light-harvesting complex II homotrimer |
|---|---|
| Components |
|
-Supramolecule #1: Bryopsis corticulans chlorophyll a/b-type light-harvesting comple...
| Supramolecule | Name: Bryopsis corticulans chlorophyll a/b-type light-harvesting complex II homotrimer type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Bryopsis corticulans (plant) Bryopsis corticulans (plant) |
-Macromolecule #1: Siphonaxanthin chlorophyll a/b binding light-harvesting complex I...
| Macromolecule | Name: Siphonaxanthin chlorophyll a/b binding light-harvesting complex II, Bry-Lhcb1 type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Bryopsis corticulans (plant) Bryopsis corticulans (plant) |
| Molecular weight | Theoretical: 26.696133 KDa |
| Sequence | String: MASAMIAQTV VGAQAARATK QVTRAAVEFY GPDRALWLGP YSEGAVPSYL TGEFPGDYGW DSAGLSADPE TFAANRELEL IHARWAMLG TVGCLTPEAL EKYGGVEFGE AVWFKAGSQI FAEGGLDYLG NPSLVHAQSI LAICWTQVVL MGLAEGYRCS G GPLGEATD ...String: MASAMIAQTV VGAQAARATK QVTRAAVEFY GPDRALWLGP YSEGAVPSYL TGEFPGDYGW DSAGLSADPE TFAANRELEL IHARWAMLG TVGCLTPEAL EKYGGVEFGE AVWFKAGSQI FAEGGLDYLG NPSLVHAQSI LAICWTQVVL MGLAEGYRCS G GPLGEATD PLYPGEAFDP MGMADDPETF AELKTKEIKN GRLAMFAMFG FFVQSLQTGK GPVECWAEHI ADPVANNGFV YA TKFFGAQ IF |
-Macromolecule #2: Siphonein
| Macromolecule | Name: Siphonein / type: ligand / ID: 2 / Number of copies: 3 / Formula: 0UR |
|---|---|
| Molecular weight | Theoretical: 781.157 Da |
| Chemical component information |  ChemComp-0UR: |
-Macromolecule #3: Siphonaxanthin
| Macromolecule | Name: Siphonaxanthin / type: ligand / ID: 3 / Number of copies: 6 / Formula: 0IE |
|---|---|
| Molecular weight | Theoretical: 600.87 Da |
| Chemical component information |  ChemComp-0IE: |
-Macromolecule #4: (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY...
| Macromolecule | Name: (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL type: ligand / ID: 4 / Number of copies: 3 / Formula: NEX |
|---|---|
| Molecular weight | Theoretical: 600.87 Da |
| Chemical component information |  ChemComp-NEX: |
-Macromolecule #5: CHLOROPHYLL B
| Macromolecule | Name: CHLOROPHYLL B / type: ligand / ID: 5 / Number of copies: 24 / Formula: CHL |
|---|---|
| Molecular weight | Theoretical: 907.472 Da |
| Chemical component information |  ChemComp-CHL: |
-Macromolecule #6: CHLOROPHYLL A
| Macromolecule | Name: CHLOROPHYLL A / type: ligand / ID: 6 / Number of copies: 18 / Formula: CLA |
|---|---|
| Molecular weight | Theoretical: 893.489 Da |
| Chemical component information |  ChemComp-CLA: |
-Macromolecule #7: 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE
| Macromolecule | Name: 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE / type: ligand / ID: 7 / Number of copies: 3 / Formula: LHG |
|---|---|
| Molecular weight | Theoretical: 722.97 Da |
| Chemical component information |  ChemComp-LHG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)