+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | dsRNA transporter | |||||||||
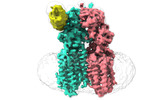
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | dsRNA transporter / RNA BINDING PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationRNA transmembrane transporter activity / dsRNA transport / RNA transport / regulatory ncRNA-mediated post-transcriptional gene silencing / double-stranded RNA binding / lysosome / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.71 Å | |||||||||
 Authors Authors | Jiang DH / Zhang JT | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2024 Journal: Nat Struct Mol Biol / Year: 2024Title: Structural insights into double-stranded RNA recognition and transport by SID-1. Authors: Jiangtao Zhang / Chunhua Zhan / Junping Fan / Dian Wu / Ruixue Zhang / Di Wu / Xinyao Chen / Ying Lu / Ming Li / Min Lin / Jianke Gong / Daohua Jiang /  Abstract: RNA uptake by cells is critical for RNA-mediated gene interference (RNAi) and RNA-based therapeutics. In Caenorhabditis elegans, RNAi is systemic as a result of SID-1-mediated double-stranded RNA ...RNA uptake by cells is critical for RNA-mediated gene interference (RNAi) and RNA-based therapeutics. In Caenorhabditis elegans, RNAi is systemic as a result of SID-1-mediated double-stranded RNA (dsRNA) across cells. Despite the functional importance, the underlying mechanisms of dsRNA internalization by SID-1 remain elusive. Here we describe cryogenic electron microscopy structures of SID-1, SID-1-dsRNA complex and human SID-1 homologs SIDT1 and SIDT2, elucidating the structural basis of dsRNA recognition and import by SID-1. The homodimeric SID-1 homologs share conserved architecture, but only SID-1 possesses the molecular determinants within its extracellular domains for distinguishing dsRNA from single-stranded RNA and DNA. We show that the removal of the long intracellular loop between transmembrane helix 1 and 2 attenuates dsRNA uptake and systemic RNAi in vivo, suggesting a possible endocytic mechanism of SID-1-mediated dsRNA internalization. Our study provides mechanistic insights into dsRNA internalization by SID-1, which may facilitate the development of dsRNA applications based on SID-1. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_34850.map.gz emd_34850.map.gz | 59.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-34850-v30.xml emd-34850-v30.xml emd-34850.xml emd-34850.xml | 15.3 KB 15.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_34850.png emd_34850.png | 76.5 KB | ||
| Filedesc metadata |  emd-34850.cif.gz emd-34850.cif.gz | 5.7 KB | ||
| Others |  emd_34850_half_map_1.map.gz emd_34850_half_map_1.map.gz emd_34850_half_map_2.map.gz emd_34850_half_map_2.map.gz | 59.4 MB 59.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-34850 http://ftp.pdbj.org/pub/emdb/structures/EMD-34850 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34850 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34850 | HTTPS FTP |
-Validation report
| Summary document |  emd_34850_validation.pdf.gz emd_34850_validation.pdf.gz | 868.2 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_34850_full_validation.pdf.gz emd_34850_full_validation.pdf.gz | 867.8 KB | Display | |
| Data in XML |  emd_34850_validation.xml.gz emd_34850_validation.xml.gz | 12.2 KB | Display | |
| Data in CIF |  emd_34850_validation.cif.gz emd_34850_validation.cif.gz | 14.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34850 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34850 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34850 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34850 | HTTPS FTP |
-Related structure data
| Related structure data |  8hkeMC  8hipC  8j6mC  8j6oC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_34850.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_34850.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.04 Å | ||||||||||||||||||||||||||||||||||||
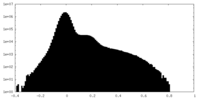
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_34850_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
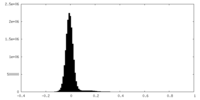
| Density Histograms |
-Half map: #1
| File | emd_34850_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
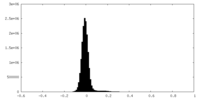
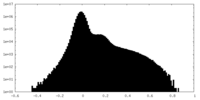
| Density Histograms |
- Sample components
Sample components
-Entire : dsRNA transporter
| Entire | Name: dsRNA transporter |
|---|---|
| Components |
|
-Supramolecule #1: dsRNA transporter
| Supramolecule | Name: dsRNA transporter / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Systemic RNA interference defective protein 1
| Macromolecule | Name: Systemic RNA interference defective protein 1 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 88.02757 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MIRVYLIILM HLVIGLTQNN STTPSPIITS SNSSVLVFEI SSKMKMIEKK LEANTVHVLR LELDQSFILD LTKVAAEIVD SSKYSKEDG VILEVTVSNG RDSFLLKLPT VYPNLKLYTD GKLLNPLVEQ DFGAHRKRHR IGDPHFHQNL IVTVQSRLNA D IDYRLHVT ...String: MIRVYLIILM HLVIGLTQNN STTPSPIITS SNSSVLVFEI SSKMKMIEKK LEANTVHVLR LELDQSFILD LTKVAAEIVD SSKYSKEDG VILEVTVSNG RDSFLLKLPT VYPNLKLYTD GKLLNPLVEQ DFGAHRKRHR IGDPHFHQNL IVTVQSRLNA D IDYRLHVT HLDRAQYDFL KFKTGQTTKT LSNQKLTFVK PIGFFLNCSE QNISQFHVTL YSEDDICANL ITVPANESIY DR SVISDKT HNRRVLSFTK RADIFFTETE ISMFKSFRIF VFIAPDDSGC STNTSRKSFN EKKKISFEFK KLENQSYAVP TAL MMIFLT TPCLLFLPIV INIIKNSRKL APSQSNLISF SPVPSEQRDM DLSHDEQQNT SSELENNGEI PAAENQIVEE ITAE NQETS VEEGNREIQV KIPLKQDSLS LHGQMLQYPV AIILPVLMHT AIEFHKWTTS TMANRDEMCF HNHACARPLG ELRAW NNII TNIGYTLYGA IFIVLSICRR GRHEYSHVFG TYECTLLDVT IGVFMVLQSI ASATYHICPS DVAFQFDTPC IQVICG LLM VRQWFVRHES PSPAYTNILL VGVVSLNFLI SAFSKTSYVR FIIAVIHVIV VGSICLAKER SLGSEKLKTR FFIMAFS MG NFAAIVMYLT LSAFHLNQIA TYCFIINCIM YLMYYGCMKV LHSERITSKA KLCGALSLLA WAVAGFFFFQ DDTDWTRS A AASRALNKPC LLLGFFGSHD LWHIFGALAG LFTFIFVSFV DDDLINTRKT SINIF UniProtKB: Systemic RNA interference defective protein 1 |
-Macromolecule #2: RNA (37-MER)
| Macromolecule | Name: RNA (37-MER) / type: rna / ID: 2 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 11.894137 KDa |
| Sequence | String: GGGCAAUGUG ACUGCAUCAG CAGUCACAUU GCCCAAG |
-Macromolecule #3: RNA (37-MER)
| Macromolecule | Name: RNA (37-MER) / type: rna / ID: 3 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 11.825017 KDa |
| Sequence | String: CUUGGGCAAU GUGACUGCUG AUGCAGUCAC AUUGCCC |
-Macromolecule #4: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 4 / Number of copies: 2 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.2 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: OTHER |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.71 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 108090 |
| Initial angle assignment | Type: RANDOM ASSIGNMENT |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)