+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the Cas12m2-crRNA binary complex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | CRISPR-Cas / RNA BINDING PROTEIN-DNA COMPLEX / RNA BINDING PROTEIN | |||||||||
| Biological species |  Mycolicibacterium mucogenicum (bacteria) Mycolicibacterium mucogenicum (bacteria) | |||||||||
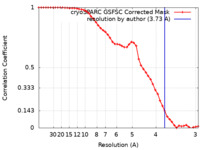
| Method | single particle reconstruction / cryo EM / Resolution: 3.73 Å | |||||||||
 Authors Authors | Omura NS / Nakagawa R / Wu YW / Sudfeld C / Warren VR / Hirano H / Kusakizako T / Kise Y / Lebbink HGJ / Itoh Y ...Omura NS / Nakagawa R / Wu YW / Sudfeld C / Warren VR / Hirano H / Kusakizako T / Kise Y / Lebbink HGJ / Itoh Y / Oost VDJ / Nureki O | |||||||||
| Funding support |  Japan, 1 items Japan, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2023 Journal: Nat Struct Mol Biol / Year: 2023Title: Mechanistic and evolutionary insights into a type V-M CRISPR-Cas effector enzyme. Authors: Satoshi N Omura / Ryoya Nakagawa / Christian Südfeld / Ricardo Villegas Warren / Wen Y Wu / Hisato Hirano / Charlie Laffeber / Tsukasa Kusakizako / Yoshiaki Kise / Joyce H G Lebbink / ...Authors: Satoshi N Omura / Ryoya Nakagawa / Christian Südfeld / Ricardo Villegas Warren / Wen Y Wu / Hisato Hirano / Charlie Laffeber / Tsukasa Kusakizako / Yoshiaki Kise / Joyce H G Lebbink / Yuzuru Itoh / John van der Oost / Osamu Nureki /   Abstract: RNA-guided type V CRISPR-Cas12 effectors provide adaptive immunity against mobile genetic elements (MGEs) in bacteria and archaea. Among diverse Cas12 enzymes, the recently identified Cas12m2 (CRISPR- ...RNA-guided type V CRISPR-Cas12 effectors provide adaptive immunity against mobile genetic elements (MGEs) in bacteria and archaea. Among diverse Cas12 enzymes, the recently identified Cas12m2 (CRISPR-Cas type V-M) is highly compact and has a unique RuvC active site. Although the non-canonical RuvC triad does not permit dsDNA cleavage, Cas12m2 still protects against invading MGEs through transcriptional silencing by strong DNA binding. However, the molecular mechanism of RNA-guided genome inactivation by Cas12m2 remains unknown. Here we report cryo-electron microscopy structures of two states of Cas12m2-CRISPR RNA (crRNA)-target DNA ternary complexes and the Cas12m2-crRNA binary complex, revealing structural dynamics during crRNA-target DNA heteroduplex formation. The structures indicate that the non-target DNA strand is tightly bound to a unique arginine-rich cluster in the recognition (REC) domains and the non-canonical active site in the RuvC domain, ensuring strong DNA-binding affinity of Cas12m2. Furthermore, a structural comparison of Cas12m2 with TnpB, a putative ancestor of Cas12 enzymes, suggests that the interaction of the characteristic coiled-coil REC2 insertion with the protospacer-adjacent motif-distal region of the heteroduplex is crucial for Cas12m2 to engage in adaptive immunity. Collectively, our findings improve mechanistic understanding of diverse type V CRISPR-Cas effectors and provide insights into the evolution of TnpB to Cas12 enzymes. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_34824.map.gz emd_34824.map.gz | 3.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-34824-v30.xml emd-34824-v30.xml emd-34824.xml emd-34824.xml | 16.3 KB 16.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_34824_fsc.xml emd_34824_fsc.xml | 4.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_34824.png emd_34824.png | 84.3 KB | ||
| Masks |  emd_34824_msk_1.map emd_34824_msk_1.map | 3.5 MB |  Mask map Mask map | |
| Others |  emd_34824_half_map_1.map.gz emd_34824_half_map_1.map.gz emd_34824_half_map_2.map.gz emd_34824_half_map_2.map.gz | 3.2 MB 3.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-34824 http://ftp.pdbj.org/pub/emdb/structures/EMD-34824 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34824 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34824 | HTTPS FTP |
-Validation report
| Summary document |  emd_34824_validation.pdf.gz emd_34824_validation.pdf.gz | 678.6 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_34824_full_validation.pdf.gz emd_34824_full_validation.pdf.gz | 678.2 KB | Display | |
| Data in XML |  emd_34824_validation.xml.gz emd_34824_validation.xml.gz | 8.9 KB | Display | |
| Data in CIF |  emd_34824_validation.cif.gz emd_34824_validation.cif.gz | 11.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34824 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34824 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34824 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34824 | HTTPS FTP |
-Related structure data
| Related structure data |  8hioMC  8hhlC  8hhmC M: atomic model generated by this map C: citing same article ( |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_34824.map.gz / Format: CCP4 / Size: 3.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_34824.map.gz / Format: CCP4 / Size: 3.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.4229 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_34824_msk_1.map emd_34824_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
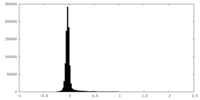
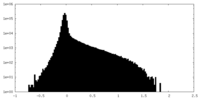
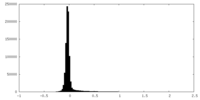
| Density Histograms |
-Half map: #1
| File | emd_34824_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_34824_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Cas12m2-crRNA
| Entire | Name: Cas12m2-crRNA |
|---|---|
| Components |
|
-Supramolecule #1: Cas12m2-crRNA
| Supramolecule | Name: Cas12m2-crRNA / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  Mycolicibacterium mucogenicum (bacteria) Mycolicibacterium mucogenicum (bacteria) |
-Supramolecule #2: crRNA
| Supramolecule | Name: crRNA / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|
-Supramolecule #3: Cas12m2
| Supramolecule | Name: Cas12m2 / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2 |
|---|
-Macromolecule #1: RNA (56-MER)
| Macromolecule | Name: RNA (56-MER) / type: rna / ID: 1 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  Mycolicibacterium mucogenicum (bacteria) Mycolicibacterium mucogenicum (bacteria) |
| Molecular weight | Theoretical: 18.277932 KDa |
| Sequence | String: GUGUCAUAGC CCAGCUUGGC GGGCGAAGGC CAAGACGGAG AUGAGGUGCG CGUGGC |
-Macromolecule #2: Cas12m2
| Macromolecule | Name: Cas12m2 / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Mycolicibacterium mucogenicum (bacteria) Mycolicibacterium mucogenicum (bacteria) |
| Molecular weight | Theoretical: 66.342883 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MTTMTVHTMG VHYKWQIPEV LRQQLWLAHN LREDLVSLQL AYDDDLKAIW SSYPDVAQAE DTMAAAEADA VALSERVKQA RIEARSKKI STELTQQLRD AKKRLKDARQ ARRDAIAVVK DDAAERRKAR SDQLAADQKA LYGQYCRDGD LYWASFNTVL D HHKTAVKR ...String: MTTMTVHTMG VHYKWQIPEV LRQQLWLAHN LREDLVSLQL AYDDDLKAIW SSYPDVAQAE DTMAAAEADA VALSERVKQA RIEARSKKI STELTQQLRD AKKRLKDARQ ARRDAIAVVK DDAAERRKAR SDQLAADQKA LYGQYCRDGD LYWASFNTVL D HHKTAVKR IAAQRASGKP ATLRHHRFDG SGTIAVQLQR QAGAPPRTPM VLADEAGKYR NVLHIPGWTD PDVWEQMTRS QC RQSGRVT VRMRCGSTDG QPQWIDLPVQ VHRWLPADAD ITGAELVVTR VAGIYRAKLC VTARIGDTEP VTSGPTVALH LGW RSTEEG TAVATWRSDA PLDIPFGLRT VMRVDAAGTS GIIVVPATIE RRLTRTENIA SSRSLALDAL RDKVVGWLSD NDAP TYRDA PLEAATVKQW KSPQRFASLA HAWKDNGTEI SDILWAWFSL DRKQWAQQEN GRRKALGHRD DLYRQIAAVI SDQAG HVLV DDTSVAELSA RAMERTELPT EVQQKIDRRR DHAAPGGLRA SVVAAMTRDG VPVTIVAAAD FTRTHSRCGH VNPADD RYL SNPVRCDGCG AMYDQDRSFV TLMLRAATAP SNP |
-Macromolecule #3: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 3 / Number of copies: 1 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Macromolecule #4: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 4 / Number of copies: 1 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.6 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 55.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.6 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)