+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of WeiTsing | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Pentamer / Channel / MEMBRANE PROTEIN | |||||||||
| Function / homology | membrane / Transmembrane protein Function and homology information Function and homology information | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.14 Å | |||||||||
 Authors Authors | Qin L / Tang LH / Chen YH | |||||||||
| Funding support |  China, 2 items China, 2 items
| |||||||||
 Citation Citation |  Journal: Cell / Year: 2023 Journal: Cell / Year: 2023Title: WeiTsing, a pericycle-expressed ion channel, safeguards the stele to confer clubroot resistance. Authors: Wei Wang / Li Qin / Wenjing Zhang / Linghui Tang / Chao Zhang / Xiaojing Dong / Pei Miao / Meng Shen / Huilong Du / Hangyuan Cheng / Ke Wang / Xiangyun Zhang / Min Su / Hongwei Lu / Chang Li ...Authors: Wei Wang / Li Qin / Wenjing Zhang / Linghui Tang / Chao Zhang / Xiaojing Dong / Pei Miao / Meng Shen / Huilong Du / Hangyuan Cheng / Ke Wang / Xiangyun Zhang / Min Su / Hongwei Lu / Chang Li / Qiang Gao / Xiaojuan Zhang / Yun Huang / Chengzhi Liang / Jian-Min Zhou / Yu-Hang Chen /  Abstract: Plant roots encounter numerous pathogenic microbes that often cause devastating diseases. One such pathogen, Plasmodiophora brassicae (Pb), causes clubroot disease and severe yield losses on ...Plant roots encounter numerous pathogenic microbes that often cause devastating diseases. One such pathogen, Plasmodiophora brassicae (Pb), causes clubroot disease and severe yield losses on cruciferous crops worldwide. Here, we report the isolation and characterization of WeiTsing (WTS), a broad-spectrum clubroot resistance gene from Arabidopsis. WTS is transcriptionally activated in the pericycle upon Pb infection to prevent pathogen colonization in the stele. Brassica napus carrying the WTS transgene displayed strong resistance to Pb. WTS encodes a small protein localized in the endoplasmic reticulum (ER), and its expression in plants induces immune responses. The cryoelectron microscopy (cryo-EM) structure of WTS revealed a previously unknown pentameric architecture with a central pore. Electrophysiology analyses demonstrated that WTS is a calcium-permeable cation-selective channel. Structure-guided mutagenesis indicated that channel activity is strictly required for triggering defenses. The findings uncover an ion channel analogous to resistosomes that triggers immune signaling in the pericycle. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_34710.map.gz emd_34710.map.gz | 30.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-34710-v30.xml emd-34710-v30.xml emd-34710.xml emd-34710.xml | 14.9 KB 14.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_34710.png emd_34710.png | 48.3 KB | ||
| Filedesc metadata |  emd-34710.cif.gz emd-34710.cif.gz | 5.3 KB | ||
| Others |  emd_34710_half_map_1.map.gz emd_34710_half_map_1.map.gz emd_34710_half_map_2.map.gz emd_34710_half_map_2.map.gz | 45.5 MB 48.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-34710 http://ftp.pdbj.org/pub/emdb/structures/EMD-34710 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34710 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34710 | HTTPS FTP |
-Related structure data
| Related structure data |  8hf2MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_34710.map.gz / Format: CCP4 / Size: 56.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_34710.map.gz / Format: CCP4 / Size: 56.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 1.06 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_34710_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
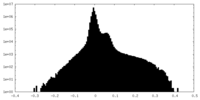
| Projections & Slices |
| ||||||||||||
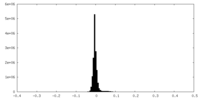
| Density Histograms |
-Half map: #2
| File | emd_34710_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
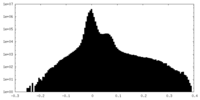
| Projections & Slices |
| ||||||||||||
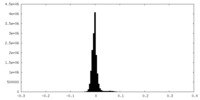
| Density Histograms |
- Sample components
Sample components
-Entire : WeiTsing
| Entire | Name: WeiTsing |
|---|---|
| Components |
|
-Supramolecule #1: WeiTsing
| Supramolecule | Name: WeiTsing / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: PRA1 family protein
| Macromolecule | Name: PRA1 family protein / type: protein_or_peptide / ID: 1 / Number of copies: 5 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 16.00399 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: METVSAVNQT LPISGGEPVK FTTYSAAVHK VLVMVNAGIL GLLQLVSQQS SVLETHKAAF LCFCVFILFY AVLRVREAMD VRLQPGLVP RLIGHGSHLF GGLAALVLVS VVSTAFSIVL FLLWFIWLSA VVYLETNKPS ACPPQLPPV UniProtKB: Transmembrane protein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2.5 mg/mL |
|---|---|
| Buffer | pH: 8 |
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: INSILICO MODEL |
|---|---|
| Final reconstruction | Applied symmetry - Point group: C5 (5 fold cyclic) / Resolution.type: BY AUTHOR / Resolution: 4.14 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: cryoSPARC (ver. 3.2) / Number images used: 117360 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller




 Z
Z Y
Y X
X