+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | IgM-var2CSA complex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | var2CSA / IgM / Placental malaria / P.falciparum / CELL ADHESION / CELL ADHESION-IMMUNE SYSTEM complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationhexameric IgM immunoglobulin complex / IgM B cell receptor complex / dimeric IgA immunoglobulin complex / secretory dimeric IgA immunoglobulin complex / pentameric IgM immunoglobulin complex / monomeric IgA immunoglobulin complex / secretory IgA immunoglobulin complex / IgA binding / pre-B cell allelic exclusion / IgM immunoglobulin complex ...hexameric IgM immunoglobulin complex / IgM B cell receptor complex / dimeric IgA immunoglobulin complex / secretory dimeric IgA immunoglobulin complex / pentameric IgM immunoglobulin complex / monomeric IgA immunoglobulin complex / secretory IgA immunoglobulin complex / IgA binding / pre-B cell allelic exclusion / IgM immunoglobulin complex / glomerular filtration / CD22 mediated BCR regulation / immunoglobulin complex, circulating / immunoglobulin receptor binding / positive regulation of respiratory burst / humoral immune response / Scavenging of heme from plasma / complement activation, classical pathway / antigen binding / Antigen activates B Cell Receptor (BCR) leading to generation of second messengers / Cell surface interactions at the vascular wall / B cell receptor signaling pathway / antibacterial humoral response / protein-macromolecule adaptor activity / protein-containing complex assembly / defense response to Gram-negative bacterium / blood microparticle / adaptive immune response / Potential therapeutics for SARS / immune response / innate immune response / cell surface / protein homodimerization activity / extracellular space / extracellular exosome / extracellular region / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /   | |||||||||
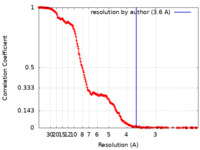
| Method | single particle reconstruction / cryo EM / Resolution: 3.6 Å | |||||||||
 Authors Authors | Akhouri RR / Goel S / Skoglund U | |||||||||
| Funding support |  Japan, 1 items Japan, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Cryo-electron microscopy of IgM-VAR2CSA complex reveals IgM inhibits binding of Plasmodium falciparum to Chondroitin Sulfate A. Authors: Reetesh Raj Akhouri / Suchi Goel / Ulf Skoglund /   Abstract: Placental malaria is caused by Plasmodium falciparum-infected erythrocytes (IEs) adhering to chondroitin sulfate proteoglycans in placenta via VAR2CSA-type PfEMP1. Human pentameric immunoglobulin M ...Placental malaria is caused by Plasmodium falciparum-infected erythrocytes (IEs) adhering to chondroitin sulfate proteoglycans in placenta via VAR2CSA-type PfEMP1. Human pentameric immunoglobulin M (IgM) binds to several types of PfEMP1, including VAR2CSA via its Fc domain. Here, a 3.6 Å cryo-electron microscopy map of the IgM-VAR2CSA complex reveals that two molecules of VAR2CSA bind to the Cµ4 of IgM through their DBL3X and DBL5ε domains. The clockwise and anti-clockwise rotation of the two VAR2CSA molecules on opposite faces of IgM juxtaposes C-termini of both VAR2CSA near the J chain, where IgM creates a wall between both VAR2CSA molecules and hinders its interaction with its receptor. To support this, we show when VAR2CSA is bound to IgM, its staining on IEs as well as binding of IEs to chondroitin sulfate A in vitro is severely compromised. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_34399.map.gz emd_34399.map.gz | 484.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-34399-v30.xml emd-34399-v30.xml emd-34399.xml emd-34399.xml | 18.4 KB 18.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_34399_fsc.xml emd_34399_fsc.xml | 24.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_34399.png emd_34399.png | 40.1 KB | ||
| Masks |  emd_34399_msk_1.map emd_34399_msk_1.map | 600.7 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-34399.cif.gz emd-34399.cif.gz | 7.6 KB | ||
| Others |  emd_34399_half_map_1.map.gz emd_34399_half_map_1.map.gz emd_34399_half_map_2.map.gz emd_34399_half_map_2.map.gz | 483.4 MB 484.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-34399 http://ftp.pdbj.org/pub/emdb/structures/EMD-34399 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34399 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34399 | HTTPS FTP |
-Validation report
| Summary document |  emd_34399_validation.pdf.gz emd_34399_validation.pdf.gz | 795.9 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_34399_full_validation.pdf.gz emd_34399_full_validation.pdf.gz | 795.4 KB | Display | |
| Data in XML |  emd_34399_validation.xml.gz emd_34399_validation.xml.gz | 26.5 KB | Display | |
| Data in CIF |  emd_34399_validation.cif.gz emd_34399_validation.cif.gz | 35.8 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34399 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34399 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34399 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34399 | HTTPS FTP |
-Related structure data
| Related structure data |  8gznMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_34399.map.gz / Format: CCP4 / Size: 600.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_34399.map.gz / Format: CCP4 / Size: 600.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.1 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_34399_msk_1.map emd_34399_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_34399_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_34399_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : IgM-var2CSA complex
| Entire | Name: IgM-var2CSA complex |
|---|---|
| Components |
|
-Supramolecule #1: IgM-var2CSA complex
| Supramolecule | Name: IgM-var2CSA complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|
-Supramolecule #2: IgM, IgJ
| Supramolecule | Name: IgM, IgJ / type: complex / ID: 2 / Parent: 1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Supramolecule #3: var2CSA
| Supramolecule | Name: var2CSA / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Immunoglobulin heavy constant mu
| Macromolecule | Name: Immunoglobulin heavy constant mu / type: protein_or_peptide / ID: 1 / Details: IgM / Number of copies: 10 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 49.48952 KDa |
| Sequence | String: GSASAPTLFP LVSCENSPSD TSSVAVGCLA QDFLPDSITF SWKYKNNSDI SSTRGFPSVL RGGKYAATSQ VLLPSKDVMQ GTDEHVVCK VQHPNGNKEK NVPLPVIAEL PPKVSVFVPP RDGFFGNPRK SKLICQATGF SPRQIQVSWL REGKQVGSGV T TDQVQAEA ...String: GSASAPTLFP LVSCENSPSD TSSVAVGCLA QDFLPDSITF SWKYKNNSDI SSTRGFPSVL RGGKYAATSQ VLLPSKDVMQ GTDEHVVCK VQHPNGNKEK NVPLPVIAEL PPKVSVFVPP RDGFFGNPRK SKLICQATGF SPRQIQVSWL REGKQVGSGV T TDQVQAEA KESGPTTYKV TSTLTIKESD WLGQSMFTCR VDHRGLTFQQ NASSMCVPDQ DTAIRVFAIP PSFASIFLTK ST KLTCLVT DLTTYDSVTI SWTRQNGEAV KTHTNISESH PNATFSAVGE ASICEDDWNS GERFTCTVTH TDLPSPLKQT ISR PKGVAL HRPDVYLLPP AREQLNLRES ATITCLVTGF SPADVFVQWM QRGQPLSPEK YVTSAPMPEP QAPGRYFAHS ILTV SEEEW NTGETYTCVV AHEALPNRVT ERTVDKSTGK PTLYNVSLVM SDTAGTCY UniProtKB: Immunoglobulin heavy constant mu |
-Macromolecule #2: Erythrocyte membrane protein 1
| Macromolecule | Name: Erythrocyte membrane protein 1 / type: protein_or_peptide / ID: 2 / Details: plasmoDB-ID: PfIT_120006100 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 311.015688 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MDSTSTIANK IEEYLGAKSD DSKIDELLKA DPSEVEYYRS GGDGDYLKNN ICKITVNHSD SGKYDPCEKK LPPYDDNDQW KCQQNSSDG SGKPENICVP PRRERLCTYN LENLKFDKIR DNNAFLADVL LTARNEGEKI VQNHPDTNSS NVCNALERSF A DLADIIRG ...String: MDSTSTIANK IEEYLGAKSD DSKIDELLKA DPSEVEYYRS GGDGDYLKNN ICKITVNHSD SGKYDPCEKK LPPYDDNDQW KCQQNSSDG SGKPENICVP PRRERLCTYN LENLKFDKIR DNNAFLADVL LTARNEGEKI VQNHPDTNSS NVCNALERSF A DLADIIRG TDQWKGTNSN LEKNLKQMFA KIRENDKVLQ DKYPKDQKYT KLREAWWNAN RQKVWEVITC GARSNDLLIK RG WRTSGKS DRKKNFELCR KCGHYEKEVP TKLDYVPQFL RWLTEWIEDF YREKQNLIDD MERHREECTR EDHKSKEGTS YCS TCKDKC KKYCECVKKW KTEWENQENK YKDLYEQNKN KTSQKNTSRY DDYVKDFFEK LEANYSSLEN YIKGDPYFAE YATK LSFIL NPSDANNPSG ETANHNDEAC NCNESGISSV GQAQTSGPSS NKTCITHSSI KTNKKKECKD VKLGVRENDK DLKIC VIED TSLSGVDNCC CQDLLGILQE NCSDNKRGSS SNDSCDNKNQ DECQKKLEKV FASLTNGYKC DKCKSGTSRS KKKWIW KKS SGNEEGLQEE YANTIGLPPR TQSLYLGNLP KLENVCEDVK DINFDTKEKF LAGCLIVSFH EGKNLKKRYP QNKNSGN KE NLCKALEYSF ADYGDLIKGT SIWDNEYTKD LELNLQNNFG KLFGKYIKKN NTAEQDTSYS SLDELRESWW NTNKKYIW T AMKHGAEMNI TTCNADGSVT GSGSSCDDIP TIDLIPQYLR FLQEWVENFC EQRQAKVKDV ITNCKSCKES GNKCKTECK TKCKDECEKY KKFIEACGTA GGGIGTAGSP WSKRWDQIYK RYSKHIEDAK RNRKAGTKNC GTSSTTNAAA STDENKCVQS DIDSFFKHL IDIGLTTPSS YLSNVLDDNI CGADKAPWTT YTTYTTTEKC NKERDKSKSQ SSDTLVVVNV PSPLGNTPYR Y KYACQCKI PTNEETCDDR KEYMNQWSCG SARTMKRGYK NDNYELCKYN GVDVKPTTVR SNSSKLDGND VTFFNLFEQW NK EIQYQIE QYMTNANISC IDEKEVLDSV SDEGTPKVRG GYEDGRNNNT DQGTNCKEKC KCYKLWIEKI NDQWGKQKDN YNK FRSKQI YDANKGSQNK KVVSLSNFLF FSCWEEYIQK YFNGDWSKIK NIGSDTFEFL IKKCGNNSAH GEEIFSEKLK NAEK KCKEN ESTDTNINKS ETSCDLNATN YIRGCQSKTY DGKIFPGKGG EKQWICKDTI IHGDTNGACI PPRTQNLCVG ELWDK SYGG RSNIKNDTKE LLKEKIKNAI HKETELLYEY HDTGTAIISK NDKKGQKGKN DPNGLPKGFC HAVQRSFIDY KNMILG TSV NIYEHIGKLQ EDIKKIIEKG TPQQKDKIGG VGSSTENVNA WWKGIEREMW DAVRCAITKI NKKNNNSIFN GDECGVS PP TGNDEDQSVS WFKEWGEQFC IERLRYEQNI REACTINGKN EKKCINSKSG QGDKIQGACK RKCEKYKKYI SEKKQEWD K QKTKYENKYV GKSASDLLKE NYPECISANF DFIFNDNIEY KTYYPYGDYS SICSCEQVKY YKYNNAEKKN NKSLCYEKD NDMTWSKKYI KKLENGRSLE GVYVPPRRQQ LCLYELFPII IKNEEGMEKA KEELLETLQI VAEREAYYLW KQYNPTGKGI DDANKKACC AIRGSFYDLE DIIKGNDLVH DEYTKYIDSK LNEIFGSSNT NDIDTKRART DWWENETITN GTDRKTIRQL V WDAMQSGV RYAVEEKNEN FPLCMGVEHI GIAKPQFIRW LEEWTNEFCE KYTKYFEDMK SKCDPPKRAD TCGDNSNIEC KK ACANYTN WLNPKRIEWN GMSNYYNKIY RKSNKESEDG KDYSMIMAPT VIDYLNKRCH GEINGNYICC SCKNIGAYNT TSG TVNKKL QKKETECEEE KGPLDLMNEV LNKMDKKYSA HKMKCTEVYL EHVEEQLNEI DNAIKDYKLY PLDRCFDDQT KMKV CDLIA DAIGCKDKTK LDELDEWNDM DLRGTYNKHK GVLIPPRRRQ LCFSRIVRGP ANLRSLNEFK EEILKGAQSE GKFLG NYYK EHKDKEKALE AMKNSFYDYE DIIKGTDMLT NIEFKDIKIK LDRLLEKETN NTKKAEDWWK TNKKSIWNAM LCGYKK SGN KIIDPSWCTI PTTETPPQFL RWIKEWGTNV CIQKQEHKEY VKSKCSNVTN LGAQASESNN CTSEIKKYQE WSRKRSI QW ETISKRYKKY KRMDILKDVK EPDANTYLRE HCSKCPCGFN DMEEMNNNED NEKEAFKQIK EQVKIPAELE DVIYRIKH H EYDKGNDYIC NKYKNIHDRM KKNNGNFVTD NFVKKSWEIS NGVLIPPRRK NLFLYIDPSK ICEYKKDPKL FKDFIYWSA FTEVERLKKA YGGARAKVVH AMKYSFTDIG SIIKGDDMME KNSSDKIGKI LGDTDGQNEK RKKWWDMNKY HIWESMLCGY REAEGDTET NENCRFPDIE SVPQFLRWFQ EWSENFCDRR QKLYDKLNSE CISAECTNGS VDNSKCTHAC VNYKNYILTK K TEYEIQTN KYDNEFKNKN SNDKDAPDYL KEKCNDNKCE CLNKHIDDKN KTWKNPYETL EDTFKSKCDC PKPLPSPIKP DD LPPQADE PFLESRGPFE GKPIPNPLLG LDSTRTGHHH HHH |
-Macromolecule #3: Immunoglobulin J chain
| Macromolecule | Name: Immunoglobulin J chain / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 15.483329 KDa |
| Sequence | String: EDERIVLVDN KCKCARITSR IIRSSEDPNE DIVERNIRII VPLNNRENIS DPTSPLRTRF VYHLSDLCKK CDPTEVELDN QIVTATQSN ICDEDSATET CYTYDRNKCY TAVVPLVYGG ETKMVETALT PDACYPD UniProtKB: Immunoglobulin J chain |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Average electron dose: 51.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller



















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)