[English] 日本語
 Yorodumi
Yorodumi- EMDB-34391: Octahedral supramolecular assembly of the bicomponent gamma-hemol... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Octahedral supramolecular assembly of the bicomponent gamma-hemolysin octameric pore complexes from Staphylococcus aureus Newman. | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Super assembly / Pore-forming toxin / cryo-EM / lipid membrane / S. aureus / TOXIN | |||||||||
| Function / homology | Bi-component toxin, staphylococci / Leukocidin/Hemolysin toxin / Leukocidin/Hemolysin toxin family / Leukocidin/porin MspA superfamily / cytolysis in another organism / toxin activity / extracellular region / Gamma-hemolysin component A / Gamma-hemolysin component B Function and homology information Function and homology information | |||||||||
| Biological species |  | |||||||||
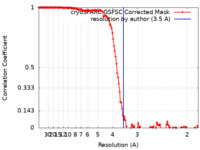
| Method | single particle reconstruction / cryo EM / Resolution: 3.5 Å | |||||||||
 Authors Authors | Mishra S / Roy A / Dutta S | |||||||||
| Funding support |  India, 2 items India, 2 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Octahedral supramolecular assembly of the bicomponent gamma-hemolysin octameric pore complexes from Staphylococcus aureus Newman. Authors: Mishra S / Roy A / Dutta S | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_34391.map.gz emd_34391.map.gz | 229.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-34391-v30.xml emd-34391-v30.xml emd-34391.xml emd-34391.xml | 14.8 KB 14.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_34391_fsc.xml emd_34391_fsc.xml | 14.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_34391.png emd_34391.png | 60.9 KB | ||
| Filedesc metadata |  emd-34391.cif.gz emd-34391.cif.gz | 5.5 KB | ||
| Others |  emd_34391_half_map_1.map.gz emd_34391_half_map_1.map.gz emd_34391_half_map_2.map.gz emd_34391_half_map_2.map.gz | 225.6 MB 225.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-34391 http://ftp.pdbj.org/pub/emdb/structures/EMD-34391 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34391 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34391 | HTTPS FTP |
-Validation report
| Summary document |  emd_34391_validation.pdf.gz emd_34391_validation.pdf.gz | 1.2 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_34391_full_validation.pdf.gz emd_34391_full_validation.pdf.gz | 1.2 MB | Display | |
| Data in XML |  emd_34391_validation.xml.gz emd_34391_validation.xml.gz | 22.4 KB | Display | |
| Data in CIF |  emd_34391_validation.cif.gz emd_34391_validation.cif.gz | 28.8 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34391 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34391 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34391 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34391 | HTTPS FTP |
-Related structure data
| Related structure data |  8gz7MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_34391.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_34391.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.92 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_34391_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_34391_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Octahedral super assembly of bicomponent Gamma-Hemolysin octameri...
| Entire | Name: Octahedral super assembly of bicomponent Gamma-Hemolysin octameric pore complexes |
|---|---|
| Components |
|
-Supramolecule #1: Octahedral super assembly of bicomponent Gamma-Hemolysin octameri...
| Supramolecule | Name: Octahedral super assembly of bicomponent Gamma-Hemolysin octameric pore complexes type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Gamma-hemolysin component B
| Macromolecule | Name: Gamma-hemolysin component B / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 31.305635 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: KVTLYKTTAT ADSDKFKISQ ILTFNFIKDK SYDKDTLVLK ATGNINSGFV KPNPNDYDFS KLYWGAKYNV SISSQSNDSV NVVDYAPKN QNEEFQVQNT LGYTFGGNTA FSETINYKQE SYRTTLSRNT NYKNVGWGVE AHKIMNNGWG PYGRDSFHPT Y GNELFLAG ...String: KVTLYKTTAT ADSDKFKISQ ILTFNFIKDK SYDKDTLVLK ATGNINSGFV KPNPNDYDFS KLYWGAKYNV SISSQSNDSV NVVDYAPKN QNEEFQVQNT LGYTFGGNTA FSETINYKQE SYRTTLSRNT NYKNVGWGVE AHKIMNNGWG PYGRDSFHPT Y GNELFLAG RQSSAYAGQN FIAQHQMPLL SRSNFNPEFL SVLSHRQDGA KKSKITVTYQ REMDLYQIRW NGFYWAGANY KN FKTRTFK STYEIDWENH KVKLLDTKET ENNK UniProtKB: Gamma-hemolysin component B |
-Macromolecule #2: Gamma-hemolysin component A
| Macromolecule | Name: Gamma-hemolysin component A / type: protein_or_peptide / ID: 2 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 30.563545 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: KIEDIGQGAE IIKRTQDITS KRLAITQNIQ FDFVKDKKYN KDALVVKMQG FISSRTTYSD LKKYPYIKRM IWPFQYNISL KTKDSNVDL INYLPKNKID SADVSQKLGY NIGSFNYSKT ISYNQKNYVT EVESQNSKGV KWGVKANSFV TPNGQVSAYD Q YLFAQDPT ...String: KIEDIGQGAE IIKRTQDITS KRLAITQNIQ FDFVKDKKYN KDALVVKMQG FISSRTTYSD LKKYPYIKRM IWPFQYNISL KTKDSNVDL INYLPKNKID SADVSQKLGY NIGSFNYSKT ISYNQKNYVT EVESQNSKGV KWGVKANSFV TPNGQVSAYD Q YLFAQDPT GPAARDYFVP DNQLPPLIQS GFNPSFITTL SHERGKGDKS EFEITYGRNM DATYAYVTRH RLAVDRKHDA FK NRNVTVK YEVNWKTHEV KIKSITPK UniProtKB: Gamma-hemolysin component A |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.8 |
|---|---|
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 293.15 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.5 µm / Nominal defocus min: 3.5 µm |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: OTHER |
|---|---|
| Output model |  PDB-8gz7: |
 Movie
Movie Controller
Controller



 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)