+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Legionella pneumophila Deubiquitinase, LotA(7-544) apo state | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Deubuiquitinase / OTU domain / SIGNALING PROTEIN | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 15.0 Å | |||||||||
 Authors Authors | Kang SW / Kim GH / Roh SH / Shin DH | |||||||||
| Funding support |  Korea, Republic Of, 2 items Korea, Republic Of, 2 items
| |||||||||
 Citation Citation |  Journal: Life Sci Alliance / Year: 2023 Journal: Life Sci Alliance / Year: 2023Title: Structural insights into ubiquitin chain cleavage by ovarian tumor deubiquitinases. Authors: Sangwoo Kang / Gyuhee Kim / Minhyeong Choi / Minwoo Jeong / Gerbrand J van der Heden van Noort / Soung-Hun Roh / Donghyuk Shin /   Abstract: Although ubiquitin is found only in eukaryotes, several pathogenic bacteria and viruses possess proteins that hinder the host ubiquitin system. , a gram-negative intracellular bacterium, possesses an ...Although ubiquitin is found only in eukaryotes, several pathogenic bacteria and viruses possess proteins that hinder the host ubiquitin system. , a gram-negative intracellular bacterium, possesses an ovarian tumor (OTU) family of deubiquitinases (Lot DUBs). Herein, we describe the molecular characteristics of Lot DUBs. We elucidated the structure of the LotA OTU1 domain and revealed that entire Lot DUBs possess a characteristic extended helical lobe that is not found in other OTU-DUBs. The structural topology of an extended helical lobe is the same throughout the Lot family, and it provides an S1' ubiquitin-binding site. Moreover, the catalytic triads of Lot DUBs resemble those of the A20-type OTU-DUBs. Furthermore, we revealed a unique mechanism by which LotA OTU domains cooperate together to distinguish the length of the chain and preferentially cleave longer K48-linked polyubiquitin chains. The LotA OTU1 domain itself cleaves K6-linked ubiquitin chains, whereas it is also essential for assisting the cleavage of longer K48-linked polyubiquitin chains by the OTU2 domain. Thus, this study provides novel insights into the structure and mechanism of action of Lot DUBs. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_34350.map.gz emd_34350.map.gz | 23.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-34350-v30.xml emd-34350-v30.xml emd-34350.xml emd-34350.xml | 12.2 KB 12.2 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_34350.png emd_34350.png | 189.8 KB | ||
| Filedesc metadata |  emd-34350.cif.gz emd-34350.cif.gz | 3.7 KB | ||
| Others |  emd_34350_half_map_1.map.gz emd_34350_half_map_1.map.gz emd_34350_half_map_2.map.gz emd_34350_half_map_2.map.gz | 5.5 MB 5.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-34350 http://ftp.pdbj.org/pub/emdb/structures/EMD-34350 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34350 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34350 | HTTPS FTP |
-Validation report
| Summary document |  emd_34350_validation.pdf.gz emd_34350_validation.pdf.gz | 502.6 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_34350_full_validation.pdf.gz emd_34350_full_validation.pdf.gz | 502.2 KB | Display | |
| Data in XML |  emd_34350_validation.xml.gz emd_34350_validation.xml.gz | 10.4 KB | Display | |
| Data in CIF |  emd_34350_validation.cif.gz emd_34350_validation.cif.gz | 12.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34350 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34350 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34350 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34350 | HTTPS FTP |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_34350.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_34350.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.68 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_34350_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
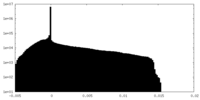
| Density Histograms |
-Half map: #2
| File | emd_34350_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
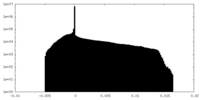
| Density Histograms |
- Sample components
Sample components
-Entire : Deubiquitinase of LotA 7-544 apo state
| Entire | Name: Deubiquitinase of LotA 7-544 apo state |
|---|---|
| Components |
|
-Supramolecule #1: Deubiquitinase of LotA 7-544 apo state
| Supramolecule | Name: Deubiquitinase of LotA 7-544 apo state / type: organelle_or_cellular_component / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.6 mg/mL |
|---|---|
| Buffer | pH: 7.6 |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV / Details: blot for 3 seconds before plunging. |
| Details | This sample was monodisperse |
- Electron microscopy
Electron microscopy
| Microscope | TFS GLACIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 60.04 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 1.8 µm / Nominal defocus min: 1.0 µm |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 15.0 Å / Resolution method: OTHER / Number images used: 176036 |
| Initial angle assignment | Type: ANGULAR RECONSTITUTION |
| Final angle assignment | Type: ANGULAR RECONSTITUTION |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)