+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | APOE4 receptor in complex with APOE4 NTD | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords |  Receptor complex / Receptor complex /  IMMUNE SYSTEM IMMUNE SYSTEM | |||||||||
| Function / homology |  Function and homology information Function and homology informationchylomicron remnant / lipid transport involved in lipid storage / triglyceride-rich lipoprotein particle clearance / intermediate-density lipoprotein particle clearance / positive regulation of lipid transport across blood-brain barrier / positive regulation of heparan sulfate proteoglycan binding / regulation of cellular response to very-low-density lipoprotein particle stimulus / metal chelating activity / Transcriptional regulation by the AP-2 (TFAP2) family of transcription factors / discoidal high-density lipoprotein particle ...chylomicron remnant / lipid transport involved in lipid storage / triglyceride-rich lipoprotein particle clearance / intermediate-density lipoprotein particle clearance / positive regulation of lipid transport across blood-brain barrier / positive regulation of heparan sulfate proteoglycan binding / regulation of cellular response to very-low-density lipoprotein particle stimulus / metal chelating activity / Transcriptional regulation by the AP-2 (TFAP2) family of transcription factors / discoidal high-density lipoprotein particle / chylomicron remnant clearance /  lipoprotein particle / maintenance of location in cell / very-low-density lipoprotein particle clearance / intermediate-density lipoprotein particle / very-low-density lipoprotein particle remodeling / regulation of amyloid-beta clearance / response to caloric restriction / Chylomicron clearance / negative regulation of triglyceride metabolic process / NMDA glutamate receptor clustering / lipoprotein particle / maintenance of location in cell / very-low-density lipoprotein particle clearance / intermediate-density lipoprotein particle / very-low-density lipoprotein particle remodeling / regulation of amyloid-beta clearance / response to caloric restriction / Chylomicron clearance / negative regulation of triglyceride metabolic process / NMDA glutamate receptor clustering /  chylomicron / Chylomicron remodeling / phosphatidylcholine-sterol O-acyltransferase activator activity / positive regulation of phospholipid efflux / chylomicron / Chylomicron remodeling / phosphatidylcholine-sterol O-acyltransferase activator activity / positive regulation of phospholipid efflux /  Chylomicron assembly / lipid transporter activity / positive regulation of low-density lipoprotein particle receptor catabolic process / positive regulation of cholesterol metabolic process / regulation of behavioral fear response / high-density lipoprotein particle remodeling / high-density lipoprotein particle clearance / Chylomicron assembly / lipid transporter activity / positive regulation of low-density lipoprotein particle receptor catabolic process / positive regulation of cholesterol metabolic process / regulation of behavioral fear response / high-density lipoprotein particle remodeling / high-density lipoprotein particle clearance /  multivesicular body, internal vesicle / lipoprotein catabolic process / very-low-density lipoprotein particle receptor binding / phospholipid efflux / regulation of amyloid fibril formation / regulation of protein metabolic process / AMPA glutamate receptor clustering / low-density lipoprotein particle / positive regulation by host of viral process / cholesterol transfer activity / multivesicular body, internal vesicle / lipoprotein catabolic process / very-low-density lipoprotein particle receptor binding / phospholipid efflux / regulation of amyloid fibril formation / regulation of protein metabolic process / AMPA glutamate receptor clustering / low-density lipoprotein particle / positive regulation by host of viral process / cholesterol transfer activity /  reverse cholesterol transport / positive regulation of amyloid-beta clearance / high-density lipoprotein particle assembly / very-low-density lipoprotein particle / positive regulation of CoA-transferase activity / melanosome organization / lipoprotein biosynthetic process / protein import / negative regulation of blood coagulation / low-density lipoprotein particle remodeling / high-density lipoprotein particle / negative regulation of amyloid fibril formation / negative regulation of cholesterol biosynthetic process / amyloid precursor protein metabolic process / reverse cholesterol transport / positive regulation of amyloid-beta clearance / high-density lipoprotein particle assembly / very-low-density lipoprotein particle / positive regulation of CoA-transferase activity / melanosome organization / lipoprotein biosynthetic process / protein import / negative regulation of blood coagulation / low-density lipoprotein particle remodeling / high-density lipoprotein particle / negative regulation of amyloid fibril formation / negative regulation of cholesterol biosynthetic process / amyloid precursor protein metabolic process /  heparan sulfate proteoglycan binding / heparan sulfate proteoglycan binding /  regulation of Cdc42 protein signal transduction / triglyceride homeostasis / regulation of amyloid precursor protein catabolic process / cholesterol catabolic process / positive regulation of membrane protein ectodomain proteolysis / regulation of Cdc42 protein signal transduction / triglyceride homeostasis / regulation of amyloid precursor protein catabolic process / cholesterol catabolic process / positive regulation of membrane protein ectodomain proteolysis /  synaptic transmission, cholinergic / HDL remodeling / negative regulation of endothelial cell migration / cholesterol efflux / Scavenging by Class A Receptors / negative regulation of protein metabolic process / artery morphogenesis / triglyceride metabolic process / regulation of axon extension / regulation of cholesterol metabolic process / positive regulation of amyloid fibril formation / low-density lipoprotein particle receptor binding / positive regulation of dendritic spine development / synaptic transmission, cholinergic / HDL remodeling / negative regulation of endothelial cell migration / cholesterol efflux / Scavenging by Class A Receptors / negative regulation of protein metabolic process / artery morphogenesis / triglyceride metabolic process / regulation of axon extension / regulation of cholesterol metabolic process / positive regulation of amyloid fibril formation / low-density lipoprotein particle receptor binding / positive regulation of dendritic spine development /  virion assembly / virion assembly /  regulation of innate immune response / regulation of neuronal synaptic plasticity / locomotory exploration behavior / regulation of innate immune response / regulation of neuronal synaptic plasticity / locomotory exploration behavior /  lipoprotein particle binding / positive regulation of endocytosis / negative regulation of amyloid-beta formation / negative regulation of endothelial cell proliferation / lipoprotein particle binding / positive regulation of endocytosis / negative regulation of amyloid-beta formation / negative regulation of endothelial cell proliferation /  antioxidant activity / cGMP-mediated signaling / response to dietary excess / negative regulation of blood vessel endothelial cell migration / negative regulation of long-term synaptic potentiation / negative regulation of platelet activation / positive regulation of cholesterol efflux / positive regulation of dendritic spine maintenance / antioxidant activity / cGMP-mediated signaling / response to dietary excess / negative regulation of blood vessel endothelial cell migration / negative regulation of long-term synaptic potentiation / negative regulation of platelet activation / positive regulation of cholesterol efflux / positive regulation of dendritic spine maintenance /  intracellular transport / regulation of protein-containing complex assembly / negative regulation of protein secretion / intracellular transport / regulation of protein-containing complex assembly / negative regulation of protein secretion /  long-term memory / fatty acid homeostasis / long-chain fatty acid transport / positive regulation of lipid biosynthetic process / long-term memory / fatty acid homeostasis / long-chain fatty acid transport / positive regulation of lipid biosynthetic process /  synaptic cleft / regulation of proteasomal protein catabolic process synaptic cleft / regulation of proteasomal protein catabolic processSimilarity search - Function | |||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.0 Å cryo EM / Resolution: 3.0 Å | |||||||||
 Authors Authors | Zhou J / Wang Y / Huang G / Shi Y | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Cell Res / Year: 2023 Journal: Cell Res / Year: 2023Title: LilrB3 is a putative cell surface receptor of APOE4. Authors: Jiayao Zhou / Yumeng Wang / Gaoxingyu Huang / Min Yang / Yumin Zhu / Chen Jin / Dan Jing / Kai Ji / Yigong Shi /  Abstract: The three isoforms of apolipoprotein E (APOE2, APOE3, and APOE4) only differ in two amino acid positions but exert quite different immunomodulatory effects. The underlying mechanism of such APOE ...The three isoforms of apolipoprotein E (APOE2, APOE3, and APOE4) only differ in two amino acid positions but exert quite different immunomodulatory effects. The underlying mechanism of such APOE isoform dependence remains enigmatic. Here we demonstrate that APOE4, but not APOE2, specifically interacts with the leukocyte immunoglobulin-like receptor B3 (LilrB3). Two discrete immunoglobin-like domains of the LilrB3 extracellular domain (ECD) recognize a positively charged surface patch on the N-terminal domain (NTD) of APOE4. The atomic structure reveals how two APOE4 molecules specifically engage two LilrB3 molecules, bringing their intracellular signaling motifs into close proximity through formation of a hetero-tetrameric complex. Consistent with our biochemical and structural analyses, APOE4, but not APOE2, activates human microglia cells (HMC3) into a pro-inflammatory state in a LilrB3-dependent manner. Together, our study identifies LilrB3 as a putative immune cell surface receptor for APOE4, but not APOE2, and may have implications for understanding the biological functions as well as disease relevance of the APOE isoforms. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_34216.map.gz emd_34216.map.gz | 59.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-34216-v30.xml emd-34216-v30.xml emd-34216.xml emd-34216.xml | 15 KB 15 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_34216.png emd_34216.png | 76.2 KB | ||
| Others |  emd_34216_half_map_1.map.gz emd_34216_half_map_1.map.gz emd_34216_half_map_2.map.gz emd_34216_half_map_2.map.gz | 59.3 MB 59.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-34216 http://ftp.pdbj.org/pub/emdb/structures/EMD-34216 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34216 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34216 | HTTPS FTP |
-Related structure data
| Related structure data |  8grxMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_34216.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_34216.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 1.0979 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_34216_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
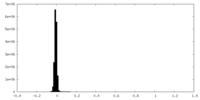
| Density Histograms |
-Half map: #1
| File | emd_34216_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
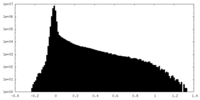
| Density Histograms |
- Sample components
Sample components
-Entire : APOE4-LilrB3 complex
| Entire | Name: APOE4-LilrB3 complex |
|---|---|
| Components |
|
-Supramolecule #1: APOE4-LilrB3 complex
| Supramolecule | Name: APOE4-LilrB3 complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 136.6 KDa |
-Macromolecule #1: Apolipoprotein E
| Macromolecule | Name: Apolipoprotein E / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 16.469791 KDa |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: GQRWELALGR FWDYLRWVQT LSEQVQEELL SSQVTQELRA LMDETMKELK AYKSELEEQL TPVAEETRAR LSKELQAAQA RLGADMEDV RGRLVQYRGE VQAMLGQSTE ELRVRLASHL RKLRKRLLRD ADDLQKRLAV Y |
-Macromolecule #2: Leukocyte immunoglobulin-like receptor subfamily A member 6
| Macromolecule | Name: Leukocyte immunoglobulin-like receptor subfamily A member 6 type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 43.869168 KDa |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: PFPKPTLWAE PGSVISWGSP VTIWCQGSLE AQEYRLDKEG SPEPLDRNNP LEPKNKARFS IPSMTEHHAG RYRCHYYSSA GWSEPSDPL ELVMTGFYNK PTLSALPSPV VASGGNMTLR CGSQKGYHHF VLMKEGEHQL PRTLDSQQLH SGGFQALFPV G PVNPSHRW ...String: PFPKPTLWAE PGSVISWGSP VTIWCQGSLE AQEYRLDKEG SPEPLDRNNP LEPKNKARFS IPSMTEHHAG RYRCHYYSSA GWSEPSDPL ELVMTGFYNK PTLSALPSPV VASGGNMTLR CGSQKGYHHF VLMKEGEHQL PRTLDSQQLH SGGFQALFPV G PVNPSHRW RFTCYYYYMN TPQVWSHPSD PLEILPSGVS RKPSLLTLQG PVLAPGQSLT LQCGSDVGYD RFVLYKEGER DF LQRPGQQ PQAGLSQANF TLGPVSRSHG GQYRCYGAHN LSSEWSAPSD PLNILMAGQI YDTVSLSAQP GPTVASGENV TLL CQSWWQ FDTFLLTKEG AAHPPLRLRS MYGAHKYQAE FPMSPVTSAH AGTYRCYGSY SSNPHLLSFP SEPLELMVSG |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 1.8 µm / Nominal defocus min: 1.5 µm Bright-field microscopy / Nominal defocus max: 1.8 µm / Nominal defocus min: 1.5 µm |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: INSILICO MODEL |
|---|---|
| Initial angle assignment | Type: NOT APPLICABLE / Software - Name: cryoSPARC |
| Final 3D classification | Software - Name: cryoSPARC |
| Final angle assignment | Type: NOT APPLICABLE / Software - Name: cryoSPARC |
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Resolution.type: BY AUTHOR / Resolution: 3.0 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 462565 |
 Movie
Movie Controller
Controller





















 Z
Z Y
Y X
X