+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | E. hirae V-ATPase state 1' (whole complex) | ||||||||||||||||||
 Map data Map data | EhVATPase S1p | ||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||
 Keywords Keywords | Sodium transport / V-ATPase / Membrane complex / MEMBRANE PROTEIN | ||||||||||||||||||
| Biological species |  Enterococcus hirae ATCC 9790 (bacteria) Enterococcus hirae ATCC 9790 (bacteria) | ||||||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 7.7 Å | ||||||||||||||||||
 Authors Authors | Burton Smith RN / Murata K | ||||||||||||||||||
| Funding support |  Japan, 5 items Japan, 5 items
| ||||||||||||||||||
 Citation Citation |  Journal: Commun Biol / Year: 2023 Journal: Commun Biol / Year: 2023Title: Six states of Enterococcus hirae V-type ATPase reveals non-uniform rotor rotation during turnover. Authors: Raymond N Burton-Smith / Chihong Song / Hiroshi Ueno / Takeshi Murata / Ryota Iino / Kazuyoshi Murata /  Abstract: The vacuolar-type ATPase from Enterococcus hirae (EhV-ATPase) is a thus-far unique adaptation of V-ATPases, as it performs Na transport and demonstrates an off-axis rotor assembly. Recent single ...The vacuolar-type ATPase from Enterococcus hirae (EhV-ATPase) is a thus-far unique adaptation of V-ATPases, as it performs Na transport and demonstrates an off-axis rotor assembly. Recent single molecule studies of the isolated V domain have indicated that there are subpauses within the three major states of the pseudo three-fold symmetric rotary enzyme. However, there was no structural evidence for these. Herein we activate the EhV-ATPase complex with ATP and identified multiple structures consisting of a total of six states of this complex by using cryo-electron microscopy. The orientations of the rotor complex during turnover, especially in the intermediates, are not as perfectly uniform as expected. The densities in the nucleotide binding pockets in the V domain indicate the different catalytic conditions for the six conformations. The off-axis rotor and its' interactions with the stator a-subunit during rotation suggests that this non-uniform rotor rotation is performed through the entire complex. | ||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_34140.map.gz emd_34140.map.gz | 17 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-34140-v30.xml emd-34140-v30.xml emd-34140.xml emd-34140.xml | 14.5 KB 14.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_34140_fsc.xml emd_34140_fsc.xml | 12.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_34140.png emd_34140.png | 73.4 KB | ||
| Others |  emd_34140_additional_1.map.gz emd_34140_additional_1.map.gz emd_34140_half_map_1.map.gz emd_34140_half_map_1.map.gz emd_34140_half_map_2.map.gz emd_34140_half_map_2.map.gz | 104.6 MB 140.5 MB 140.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-34140 http://ftp.pdbj.org/pub/emdb/structures/EMD-34140 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34140 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34140 | HTTPS FTP |
-Validation report
| Summary document |  emd_34140_validation.pdf.gz emd_34140_validation.pdf.gz | 795.3 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_34140_full_validation.pdf.gz emd_34140_full_validation.pdf.gz | 794.8 KB | Display | |
| Data in XML |  emd_34140_validation.xml.gz emd_34140_validation.xml.gz | 19.8 KB | Display | |
| Data in CIF |  emd_34140_validation.cif.gz emd_34140_validation.cif.gz | 26.1 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34140 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34140 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34140 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34140 | HTTPS FTP |
-Related structure data
| Related structure data | C: citing same article ( |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_34140.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_34140.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | EhVATPase S1p | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.01 Å | ||||||||||||||||||||||||||||||||||||
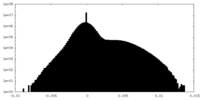
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: Locally filtered map EhVATPase S1p
| File | emd_34140_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Locally filtered map EhVATPase S1p | ||||||||||||
| Projections & Slices |
| ||||||||||||
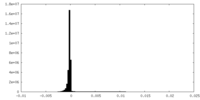
| Density Histograms |
-Half map: EhVATPase S1p half map 1
| File | emd_34140_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | EhVATPase S1p half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: EhVATPase S1p half map 2
| File | emd_34140_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | EhVATPase S1p half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : E. hirae V-ATPase state 1' (whole complex)
| Entire | Name: E. hirae V-ATPase state 1' (whole complex) |
|---|---|
| Components |
|
-Supramolecule #1: E. hirae V-ATPase state 1' (whole complex)
| Supramolecule | Name: E. hirae V-ATPase state 1' (whole complex) / type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  Enterococcus hirae ATCC 9790 (bacteria) Enterococcus hirae ATCC 9790 (bacteria) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | JEOL CRYO ARM 300 |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm |
 Movie
Movie Controller
Controller















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)