+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of VTC complex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | VTC complex / Poly P. / TRANSPORT PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationvacuolar transporter chaperone complex / ATP-polyphosphate phosphotransferase / polyphosphate biosynthetic process / engulfment of target by autophagosome / polyphosphate kinase activity / microautophagy / vacuole fusion, non-autophagic / polyphosphate metabolic process / intracellular phosphate ion homeostasis / inositol hexakisphosphate binding ...vacuolar transporter chaperone complex / ATP-polyphosphate phosphotransferase / polyphosphate biosynthetic process / engulfment of target by autophagosome / polyphosphate kinase activity / microautophagy / vacuole fusion, non-autophagic / polyphosphate metabolic process / intracellular phosphate ion homeostasis / inositol hexakisphosphate binding / vacuolar transport / fungal-type vacuole membrane / vacuolar membrane / autophagosome membrane / cell periphery / cell cortex / cytoplasmic vesicle / nuclear membrane / calmodulin binding / mRNA binding / endoplasmic reticulum membrane / endoplasmic reticulum / membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.0 Å | |||||||||
 Authors Authors | Guan ZY / Chen J / Liu RW / Chen YK / Xing Q / Du ZM / Liu Z | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: The cytoplasmic synthesis and coupled membrane translocation of eukaryotic polyphosphate by signal-activated VTC complex. Authors: Zeyuan Guan / Juan Chen / Ruiwen Liu / Yanke Chen / Qiong Xing / Zhangmeng Du / Meng Cheng / Jianjian Hu / Wenhui Zhang / Wencong Mei / Beijing Wan / Qiang Wang / Jie Zhang / Peng Cheng / ...Authors: Zeyuan Guan / Juan Chen / Ruiwen Liu / Yanke Chen / Qiong Xing / Zhangmeng Du / Meng Cheng / Jianjian Hu / Wenhui Zhang / Wencong Mei / Beijing Wan / Qiang Wang / Jie Zhang / Peng Cheng / Huanyu Cai / Jianbo Cao / Delin Zhang / Junjie Yan / Ping Yin / Michael Hothorn / Zhu Liu /   Abstract: Inorganic polyphosphate (polyP) is an ancient energy metabolite and phosphate store that occurs ubiquitously in all organisms. The vacuolar transporter chaperone (VTC) complex integrates cytosolic ...Inorganic polyphosphate (polyP) is an ancient energy metabolite and phosphate store that occurs ubiquitously in all organisms. The vacuolar transporter chaperone (VTC) complex integrates cytosolic polyP synthesis from ATP and polyP membrane translocation into the vacuolar lumen. In yeast and in other eukaryotes, polyP synthesis is regulated by inositol pyrophosphate (PP-InsP) nutrient messengers, directly sensed by the VTC complex. Here, we report the cryo-electron microscopy structure of signal-activated VTC complex at 3.0 Å resolution. Baker's yeast VTC subunits Vtc1, Vtc3, and Vtc4 assemble into a 3:1:1 complex. Fifteen trans-membrane helices form a novel membrane channel enabling the transport of newly synthesized polyP into the vacuolar lumen. PP-InsP binding orients the catalytic polymerase domain at the entrance of the trans-membrane channel, both activating the enzyme and coupling polyP synthesis and membrane translocation. Together with biochemical and cellular studies, our work provides mechanistic insights into the biogenesis of an ancient energy metabolite. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_34090.map.gz emd_34090.map.gz | 78.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-34090-v30.xml emd-34090-v30.xml emd-34090.xml emd-34090.xml | 19.1 KB 19.1 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_34090.png emd_34090.png | 91.6 KB | ||
| Filedesc metadata |  emd-34090.cif.gz emd-34090.cif.gz | 6.7 KB | ||
| Others |  emd_34090_half_map_1.map.gz emd_34090_half_map_1.map.gz emd_34090_half_map_2.map.gz emd_34090_half_map_2.map.gz | 77.9 MB 77.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-34090 http://ftp.pdbj.org/pub/emdb/structures/EMD-34090 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34090 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34090 | HTTPS FTP |
-Validation report
| Summary document |  emd_34090_validation.pdf.gz emd_34090_validation.pdf.gz | 815.6 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_34090_full_validation.pdf.gz emd_34090_full_validation.pdf.gz | 815.2 KB | Display | |
| Data in XML |  emd_34090_validation.xml.gz emd_34090_validation.xml.gz | 12.7 KB | Display | |
| Data in CIF |  emd_34090_validation.cif.gz emd_34090_validation.cif.gz | 14.6 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34090 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34090 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34090 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34090 | HTTPS FTP |
-Related structure data
| Related structure data |  7ytjMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_34090.map.gz / Format: CCP4 / Size: 83.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_34090.map.gz / Format: CCP4 / Size: 83.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 1.07 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_34090_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
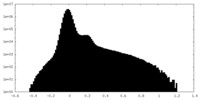
| Projections & Slices |
| ||||||||||||
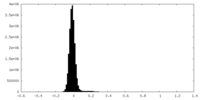
| Density Histograms |
-Half map: #2
| File | emd_34090_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
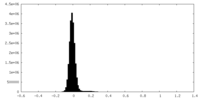
| Density Histograms |
- Sample components
Sample components
-Entire : VTC complex
| Entire | Name: VTC complex |
|---|---|
| Components |
|
-Supramolecule #1: VTC complex
| Supramolecule | Name: VTC complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Vacuolar transporter chaperone 4
| Macromolecule | Name: Vacuolar transporter chaperone 4 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 86.425984 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MAKFGEHLSK SLIRQYSYYY ISYDDLKTEL EDNLSKNNGQ WTQELETDFL ESLEIELDKV YTFCKVKHSE VFRRVKEVQE QVQHTVRLL DSNNPPTQLD FEILEEELSD IIADVHDLAK FSRLNYTGFQ KIIKKHDKKT GFILKPVFQV RLDSKPFFKE N YDELVVKI ...String: MAKFGEHLSK SLIRQYSYYY ISYDDLKTEL EDNLSKNNGQ WTQELETDFL ESLEIELDKV YTFCKVKHSE VFRRVKEVQE QVQHTVRLL DSNNPPTQLD FEILEEELSD IIADVHDLAK FSRLNYTGFQ KIIKKHDKKT GFILKPVFQV RLDSKPFFKE N YDELVVKI SQLYDIARTS GRPIKGDSSA GGKQQNFVRQ TTKYWVHPDN ITELKLIILK HLPVLVFNTN KEFEREDSAI TS IYFDNEN LDLYYGRLRK DEGAEAHALA WYGGMSTDTI FVERKTHRED WTGEKSVKAR FALKERHVND FLKGKYTVDQ VFA KMRKEG KKPMNEIENL EALASEIQYV MLKKKLRPVV RSFYNRTAFQ LPGDARVRIS LDTELTMVRE DNFDGVDRTH KNWR RTDIG VDWPFKQLDD KDICRFPYAV LNVKLQTQLG QEPPEWVREL VGSHLVEPVP KFSKFIHGVA TLLNDKVDSI PFWLP QMDV DIRKPPLPTN IEITRPGRSD NEDNDFDEDD EDDAALVAAM TNAPGNSLDI EESVGYGATS APTSNTNHVV ESANAA YYQ RKIRNAENPI SKKYYEIVAF FDHYFNGDQI SKIPKGTTFD TQIRAPPGKT ICVPVRVEPK VYFATERTYL SWLSISI LL GGVSTTLLTY GSPTAMIGSI GFFITSLAVL IRTVMVYAKR VVNIRLKRAV DYEDKIGPGM VSVFLILSIL FSFFCNLV A KLESAWSHPQ FEKGGGSGGG SGGSAWSHPQ FEK UniProtKB: Vacuolar transporter chaperone complex subunit 4 |
-Macromolecule #2: Vacuolar transporter chaperone 1
| Macromolecule | Name: Vacuolar transporter chaperone 1 / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 15.943701 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MASSAPLLQR TPGKKIALPT RVEPKVFFAN ERTFLSWLNF TVMLGGLGVG LLNFGDKIGR VSAGLFTFVA MGTMIYALVT YHWRAAAIR RRGSGPYDDR LGPTLLCFFL LVAVIINFIL RLKYNDANTK LLESAYPYDV PDYA UniProtKB: Vacuolar transporter chaperone complex subunit 1 |
-Macromolecule #3: Vacuolar transporter chaperone 3
| Macromolecule | Name: Vacuolar transporter chaperone 3 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 100.120773 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MDYKDDDDKG DYKDDDDKID YKDDDDKGSM LFGIKLANDV YPPWKDSYID YERLKKLLKE SVIHDGRSSV DSWSERNESD FVEALDKEL EKVYTFQISK YNAVLRKLDD LEENTKSAEK IQKINSEQFK NTLEECLDEA QRLDNFDRLN FTGFIKIVKK H DKLHPNYP ...String: MDYKDDDDKG DYKDDDDKID YKDDDDKGSM LFGIKLANDV YPPWKDSYID YERLKKLLKE SVIHDGRSSV DSWSERNESD FVEALDKEL EKVYTFQISK YNAVLRKLDD LEENTKSAEK IQKINSEQFK NTLEECLDEA QRLDNFDRLN FTGFIKIVKK H DKLHPNYP SVKSLLQVRL KELPFNNSEE YSPLLYRISY LYEFLRSNYD HPNTVSKSLA STSKLSHFSN LEDASFKSYK FW VHDDNIM EVKARILRHL PALVYASVPN ENDDFVDNLE SDVRVQPEAR LNIGSKSNSL SSDGNSNQDV EIGKSKSVIF PQS YDPTIT TLYFDNDFFD LYNNRLLKIS GAPTLRLRWI GKLLDKPDIF LEKRTFTENT ETGNSSFEEI RLQMKAKFIN NFIF KNDPS YKNYLINQLR ERGTQKEELE KLSRDFDNIQ NFIVEEKLQP VLRATYNRTA FQIPGDQSIR VTIDSNIMYI REDSL DKNR PIRNPENWHR DDIDSNIPNP LRFLRAGEYS KFPYSVMEIK VINQDNSQMP NYEWIKDLTN SHLVNEVPKF SLYLQG VAS LFGEDDKYVN ILPFWLPDLE TDIRKNPQEA YEEEKKTLQK QKSIHDKLDN MRRLSKISVP DGKTTERQGQ KDQNTRH VI ADLEDHESSD EEGTALPKKS AVKKGKKFKT NAAFLKILAG KNISENGNDP YSDDTDSASS FQLPPGVKKP VHLLKNAG P VKVEAKVWLA NERTFNRWLS VTTLLSVLTF SIYNSVQKAE FPQLADLLAY VYFFLTLFCG VWAYRTYLKR LTLIKGRSG KHLDAPVGPI LVAVVLIVTL VVNFSVAFKE AARRERGLVN VSSQPSLPRT LKPIQDFIFN LVGE UniProtKB: Vacuolar transporter chaperone 3 complex subunit 3 |
-Macromolecule #4: INOSITOL HEXAKISPHOSPHATE
| Macromolecule | Name: INOSITOL HEXAKISPHOSPHATE / type: ligand / ID: 4 / Number of copies: 3 / Formula: IHP |
|---|---|
| Molecular weight | Theoretical: 660.035 Da |
| Chemical component information |  ChemComp-IHP: |
-Macromolecule #5: 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE
| Macromolecule | Name: 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE / type: ligand / ID: 5 / Number of copies: 2 / Formula: PC1 |
|---|---|
| Molecular weight | Theoretical: 790.145 Da |
| Chemical component information |  ChemComp-PC1: |
-Macromolecule #6: PHOSPHATE ION
| Macromolecule | Name: PHOSPHATE ION / type: ligand / ID: 6 / Number of copies: 2 / Formula: PO4 |
|---|---|
| Molecular weight | Theoretical: 94.971 Da |
| Chemical component information |  ChemComp-PO4: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 6.5 mg/mL |
|---|---|
| Buffer | pH: 8 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 55.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.2 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller




 Z
Z Y
Y X
X