[English] 日本語
 Yorodumi
Yorodumi- EMDB-34063: Cyro-EM structure of HCMV glycoprotein B in complex with 1B03 Fab -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cyro-EM structure of HCMV glycoprotein B in complex with 1B03 Fab | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | VIRAL PROTEN / VIRAL PROTEIN-IMMUNE SYSTEM complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationhost cell endosome / host cell Golgi apparatus / symbiont entry into host cell / viral envelope / virion attachment to host cell / host cell plasma membrane / membrane Similarity search - Function | |||||||||
| Biological species |   Human betaherpesvirus 5 / Human betaherpesvirus 5 /  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.99 Å | |||||||||
 Authors Authors | Wang H / Zhu S / Liao H | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Cyro-EM structure of HCMV glycoprotein B in complex with 1B03 Fab Authors: Wu C / Wang H / Zhu S / Liao H | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_34063.map.gz emd_34063.map.gz | 12 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-34063-v30.xml emd-34063-v30.xml emd-34063.xml emd-34063.xml | 16.6 KB 16.6 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_34063.png emd_34063.png | 51.1 KB | ||
| Filedesc metadata |  emd-34063.cif.gz emd-34063.cif.gz | 5.8 KB | ||
| Others |  emd_34063_half_map_1.map.gz emd_34063_half_map_1.map.gz emd_34063_half_map_2.map.gz emd_34063_half_map_2.map.gz | 140.6 MB 140.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-34063 http://ftp.pdbj.org/pub/emdb/structures/EMD-34063 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34063 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34063 | HTTPS FTP |
-Validation report
| Summary document |  emd_34063_validation.pdf.gz emd_34063_validation.pdf.gz | 752.5 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_34063_full_validation.pdf.gz emd_34063_full_validation.pdf.gz | 752.1 KB | Display | |
| Data in XML |  emd_34063_validation.xml.gz emd_34063_validation.xml.gz | 14.9 KB | Display | |
| Data in CIF |  emd_34063_validation.cif.gz emd_34063_validation.cif.gz | 17.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34063 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34063 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34063 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34063 | HTTPS FTP |
-Related structure data
| Related structure data |  7yrnMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_34063.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_34063.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.856 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_34063_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
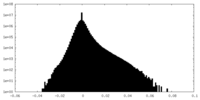
| Projections & Slices |
| ||||||||||||
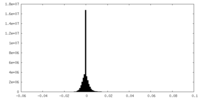
| Density Histograms |
-Half map: #2
| File | emd_34063_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
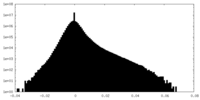
| Projections & Slices |
| ||||||||||||
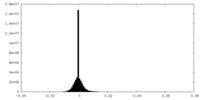
| Density Histograms |
- Sample components
Sample components
-Entire : HCMV glycoprotein B in complex with 1B03 Fab
| Entire | Name: HCMV glycoprotein B in complex with 1B03 Fab |
|---|---|
| Components |
|
-Supramolecule #1: HCMV glycoprotein B in complex with 1B03 Fab
| Supramolecule | Name: HCMV glycoprotein B in complex with 1B03 Fab / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|
-Supramolecule #2: HCMV glycoprotein B
| Supramolecule | Name: HCMV glycoprotein B / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:   Human betaherpesvirus 5 Human betaherpesvirus 5 |
-Supramolecule #3: 1B03 Fab
| Supramolecule | Name: 1B03 Fab / type: complex / ID: 3 / Parent: 2 / Macromolecule list: #2-#3 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Envelope glycoprotein B
| Macromolecule | Name: Envelope glycoprotein B / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Human betaherpesvirus 5 / Strain: Towne Human betaherpesvirus 5 / Strain: Towne |
| Molecular weight | Theoretical: 83.545547 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MESRIWCLVV CVNLCIVCLG AAVSSSSTRG TSATHSHHSS HTTSAAHSRS GSVSQRVTSS QTVSHGVNET IYNTTLKYGD VVGVNTTKY PYRVCSMAQG TDLIRFERNI VCTSMKPINE DLDEGIMVVY KRNIVAHTFK VRVYQKVLTF RRSYAYHRTT Y LLGSNTEY ...String: MESRIWCLVV CVNLCIVCLG AAVSSSSTRG TSATHSHHSS HTTSAAHSRS GSVSQRVTSS QTVSHGVNET IYNTTLKYGD VVGVNTTKY PYRVCSMAQG TDLIRFERNI VCTSMKPINE DLDEGIMVVY KRNIVAHTFK VRVYQKVLTF RRSYAYHRTT Y LLGSNTEY VAPPMWEIHH INSHSQCYSS YSRVIAGTVF VAYHRDSYEN KTMQLMPDDY SNTHSTRYVT VKDQWHSRGS TN LTRETSN LNCMVTITTA RSKYPYHFFA TSTGDVVDIS PFYNGTNRNA SYFGENADKF FIFPNYTIVS DFGRPNSALE THR LVAFLE RADSVISWDI QDEKNVTCQL TFWEASERTI RSEAEDSYHF SSAKMTATFL SKKQEVNMSD SALDCVRDEA INKL QQIFN TSYNQTYEKY GNVSVFETTG GLVVFWQGIK QKSLVELERL ANRSSLNLTH NETKESTDGN NATHLSNMES VHNLV YAQL QFTYDTLRGY INRALAQIAE AWCVDQRRTL EVFKELSKIN PSAILSAIYN KPIAARFMGD VLGLASCVTI NQTSVK VLR DMNVKESPGR CYSRPVVIFN FANSSYVQYG QLGEDNEILL GNHRTEECQL PSLKIFIAGN SAYEYVDYLF KRMIDLS SI STVDSMIALD IDPLENTDFR VLELYSQKEL RSINVFDLEE IMREFNSYKQ RVKYVEDKGL NDIFEAQKIE WHELEVLF Q GPGHHHHHHH UniProtKB: Envelope glycoprotein B |
-Macromolecule #2: 1B03 Fab antibody Heavy Chain
| Macromolecule | Name: 1B03 Fab antibody Heavy Chain / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 23.851783 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QVQLVQSGAE VKKPGESLKI SCKGSGYTFT NYWIGWVRQM PGAGLEWMAI IFPRDSYSAY SPSFQGRVTI SVDKSISTAY LHWSSLEAS DTAVYYCAIY NDLRSGNSWG QGTPLIVSSA STKGPSVFPL APSSKSTSGG TAALGCLVKD YFPEPVTVSW N SGALTSGV ...String: QVQLVQSGAE VKKPGESLKI SCKGSGYTFT NYWIGWVRQM PGAGLEWMAI IFPRDSYSAY SPSFQGRVTI SVDKSISTAY LHWSSLEAS DTAVYYCAIY NDLRSGNSWG QGTPLIVSSA STKGPSVFPL APSSKSTSGG TAALGCLVKD YFPEPVTVSW N SGALTSGV HTFPAVLQSS GLYSLSSVVT VPSSSLGTQT YICNVNHKPS NTKVDKRVEP KSCDK |
-Macromolecule #3: 1B03 Fab antibody Light Chain
| Macromolecule | Name: 1B03 Fab antibody Light Chain / type: protein_or_peptide / ID: 3 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 23.508117 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: DIQMTQSPSS LSASVGDRVT ITCRASQSIT KYLNWYQQKP GRAPKLLIHT TSTLQSGVPS RFSGSGSGTD FTLTISSLQL EDFGTYYCQ QSFSTLWTFG QGTKLDIKRT VAAPSVFIFP PSDEQLKSGT ASVVCLLNNF YPREAKVQWK VDNALQSGNS Q ESVTEQDS ...String: DIQMTQSPSS LSASVGDRVT ITCRASQSIT KYLNWYQQKP GRAPKLLIHT TSTLQSGVPS RFSGSGSGTD FTLTISSLQL EDFGTYYCQ QSFSTLWTFG QGTKLDIKRT VAAPSVFIFP PSDEQLKSGT ASVVCLLNNF YPREAKVQWK VDNALQSGNS Q ESVTEQDS KDSTYSLSST LTLSKADYEK HKVYACEVTH QGLSSPVTKS FNRGEC |
-Macromolecule #5: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 5 / Number of copies: 6 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 2.99 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 185463 |
| Initial angle assignment | Type: ANGULAR RECONSTITUTION |
| Final angle assignment | Type: ANGULAR RECONSTITUTION |
 Movie
Movie Controller
Controller



 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)