+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of hIAPP-TF-type3 | ||||||||||||
 Map data Map data | hIAPP-WF-type3 | ||||||||||||
 Sample Sample |
| ||||||||||||
| Function / homology |  Function and homology information Function and homology information: / amylin receptor signaling pathway / Calcitonin-like ligand receptors / negative regulation of amyloid fibril formation / negative regulation of bone resorption /  eating behavior / negative regulation of osteoclast differentiation / positive regulation of protein kinase A signaling / eating behavior / negative regulation of osteoclast differentiation / positive regulation of protein kinase A signaling /  Regulation of gene expression in beta cells / negative regulation of protein-containing complex assembly ...: / amylin receptor signaling pathway / Calcitonin-like ligand receptors / negative regulation of amyloid fibril formation / negative regulation of bone resorption / Regulation of gene expression in beta cells / negative regulation of protein-containing complex assembly ...: / amylin receptor signaling pathway / Calcitonin-like ligand receptors / negative regulation of amyloid fibril formation / negative regulation of bone resorption /  eating behavior / negative regulation of osteoclast differentiation / positive regulation of protein kinase A signaling / eating behavior / negative regulation of osteoclast differentiation / positive regulation of protein kinase A signaling /  Regulation of gene expression in beta cells / negative regulation of protein-containing complex assembly / positive regulation of cAMP-mediated signaling / positive regulation of calcium-mediated signaling / Regulation of gene expression in beta cells / negative regulation of protein-containing complex assembly / positive regulation of cAMP-mediated signaling / positive regulation of calcium-mediated signaling /  bone resorption / sensory perception of pain / osteoclast differentiation / bone resorption / sensory perception of pain / osteoclast differentiation /  hormone activity / cell-cell signaling / hormone activity / cell-cell signaling /  amyloid-beta binding / G alpha (s) signalling events / positive regulation of MAPK cascade / amyloid-beta binding / G alpha (s) signalling events / positive regulation of MAPK cascade /  receptor ligand activity / positive regulation of apoptotic process / Amyloid fiber formation / receptor ligand activity / positive regulation of apoptotic process / Amyloid fiber formation /  signaling receptor binding / signaling receptor binding /  lipid binding / apoptotic process / lipid binding / apoptotic process /  signal transduction / signal transduction /  extracellular space / extracellular region / identical protein binding extracellular space / extracellular region / identical protein bindingSimilarity search - Function | ||||||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | ||||||||||||
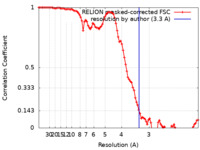
| Method | helical reconstruction / Resolution: 3.3 Å | ||||||||||||
 Authors Authors | Li D / Zhang X | ||||||||||||
| Funding support |  China, 3 items China, 3 items
| ||||||||||||
 Citation Citation |  Journal: Iscience / Year: 2022 Journal: Iscience / Year: 2022Title: A new polymorphism of human amylin fibrils with similar protofilaments and a conserved core Authors: Li D / Zhang X / Wang Y / Zhang H / Song K / Bao K / Zhu P | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_33903.map.gz emd_33903.map.gz | 3.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-33903-v30.xml emd-33903-v30.xml emd-33903.xml emd-33903.xml | 13.6 KB 13.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_33903_fsc.xml emd_33903_fsc.xml | 9.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_33903.png emd_33903.png | 67 KB | ||
| Others |  emd_33903_half_map_1.map.gz emd_33903_half_map_1.map.gz emd_33903_half_map_2.map.gz emd_33903_half_map_2.map.gz | 54.4 MB 54.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-33903 http://ftp.pdbj.org/pub/emdb/structures/EMD-33903 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33903 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33903 | HTTPS FTP |
-Related structure data
| Related structure data |  7yl7MC  7ykwC  7yl0C  7yl3C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_33903.map.gz / Format: CCP4 / Size: 70.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_33903.map.gz / Format: CCP4 / Size: 70.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | hIAPP-WF-type3 | ||||||||||||||||||||
| Voxel size | X=Y=Z: 1.04 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_33903_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_33903_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : no
| Entire | Name: no |
|---|---|
| Components |
|
-Supramolecule #1: no
| Supramolecule | Name: no / type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Islet amyloid polypeptide
| Macromolecule | Name: Islet amyloid polypeptide / type: protein_or_peptide / ID: 1 / Number of copies: 12 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 3.908319 KDa |
| Sequence | String: KCNTATCATQ RLANFLVHSS NNFGAILSST NVGSNT(TYC) |
-Experimental details
-Structure determination
 Processing Processing | helical reconstruction |
|---|---|
| Aggregation state | helical array |
- Sample preparation
Sample preparation
| Buffer | pH: 7 |
|---|
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.8 mm / Nominal defocus max: 1.5 µm / Nominal defocus min: 1.0 µm Bright-field microscopy / Cs: 2.8 mm / Nominal defocus max: 1.5 µm / Nominal defocus min: 1.0 µm |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Average exposure time: 3.0 sec. / Average electron dose: 60.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller










 Z
Z Y
Y X
X