+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Human KCNH5-closed state 2 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Potassium Channel / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationdelayed rectifier potassium channel activity / Voltage gated Potassium channels / voltage-gated potassium channel activity / voltage-gated potassium channel complex / regulation of G2/M transition of mitotic cell cycle / potassium ion transmembrane transport / regulation of membrane potential / potassium ion transport / transmembrane transporter binding / calmodulin binding ...delayed rectifier potassium channel activity / Voltage gated Potassium channels / voltage-gated potassium channel activity / voltage-gated potassium channel complex / regulation of G2/M transition of mitotic cell cycle / potassium ion transmembrane transport / regulation of membrane potential / potassium ion transport / transmembrane transporter binding / calmodulin binding / protein-containing complex binding / cell surface / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.4 Å | |||||||||
 Authors Authors | Zhang MF | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Mechanism underlying delayed rectifying in human voltage-mediated activation Eag2 channel. Authors: Mingfeng Zhang / Yuanyue Shan / Duanqing Pei /  Abstract: The transmembrane voltage gradient is a general physico-chemical cue that regulates diverse biological function through voltage-gated ion channels. How voltage sensing mediates ion flows remains ...The transmembrane voltage gradient is a general physico-chemical cue that regulates diverse biological function through voltage-gated ion channels. How voltage sensing mediates ion flows remains unknown at the molecular level. Here, we report six conformations of the human Eag2 (hEag2) ranging from closed, pre-open, open, and pore dilation but non-conducting states captured by cryo-electron microscopy (cryo-EM). These multiple states illuminate dynamics of the selectivity filter and ion permeation pathway with delayed rectifier properties and Cole-Moore effect at the atomic level. Mechanistically, a short S4-S5 linker is coupled with the constrict sites to mediate voltage transducing in a non-domain-swapped configuration, resulting transitions for constrict sites of F464 and Q472 from gating to open state stabilizing for voltage energy transduction. Meanwhile, an additional potassium ion occupied at positions S6 confers the delayed rectifier property and Cole-Moore effects. These results provide insight into voltage transducing and potassium current across membrane, and shed light on the long-sought Cole-Moore effects. | |||||||||
| History |
|
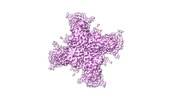
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_33856.map.gz emd_33856.map.gz | 230 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-33856-v30.xml emd-33856-v30.xml emd-33856.xml emd-33856.xml | 15.9 KB 15.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_33856.png emd_33856.png | 385.5 KB | ||
| Filedesc metadata |  emd-33856.cif.gz emd-33856.cif.gz | 6.1 KB | ||
| Others |  emd_33856_half_map_1.map.gz emd_33856_half_map_1.map.gz emd_33856_half_map_2.map.gz emd_33856_half_map_2.map.gz | 225.9 MB 225.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-33856 http://ftp.pdbj.org/pub/emdb/structures/EMD-33856 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33856 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33856 | HTTPS FTP |
-Validation report
| Summary document |  emd_33856_validation.pdf.gz emd_33856_validation.pdf.gz | 991.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_33856_full_validation.pdf.gz emd_33856_full_validation.pdf.gz | 991.4 KB | Display | |
| Data in XML |  emd_33856_validation.xml.gz emd_33856_validation.xml.gz | 15.8 KB | Display | |
| Data in CIF |  emd_33856_validation.cif.gz emd_33856_validation.cif.gz | 18.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33856 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33856 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33856 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33856 | HTTPS FTP |
-Related structure data
| Related structure data |  7yieMC  7yidC  7yifC  7yigC  7yihC  7yijC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_33856.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_33856.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.849 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_33856_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_33856_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : human KCNH5
| Entire | Name: human KCNH5 |
|---|---|
| Components |
|
-Supramolecule #1: human KCNH5
| Supramolecule | Name: human KCNH5 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 60 KDa |
-Macromolecule #1: Potassium voltage-gated channel subfamily H member 5
| Macromolecule | Name: Potassium voltage-gated channel subfamily H member 5 / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 112.011516 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MPGGKRGLVA PQNTFLENIV RRSSESSFLL GNAQIVDWPV VYSNDGFCKL SGYHRADVMQ KSSTCSFMYG ELTDKKTIEK VRQTFDNYE SNCFEVLLYK KNRTPVWFYM QIAPIRNEHE KVVLFLCTFK DITLFKQPIE DDSTKGWTKF ARLTRALTNS R SVLQQLTP ...String: MPGGKRGLVA PQNTFLENIV RRSSESSFLL GNAQIVDWPV VYSNDGFCKL SGYHRADVMQ KSSTCSFMYG ELTDKKTIEK VRQTFDNYE SNCFEVLLYK KNRTPVWFYM QIAPIRNEHE KVVLFLCTFK DITLFKQPIE DDSTKGWTKF ARLTRALTNS R SVLQQLTP MNKTEVVHKH SRLAEVLQLG SDILPQYKQE APKTPPHIIL HYCAFKTTWD WVILILTFYT AIMVPYNVSF KT KQNNIAW LVLDSVVDVI FLVDIVLNFH TTFVGPGGEV ISDPKLIRMN YLKTWFVIDL LSCLPYDIIN AFENVDEGIS SLF SSLKVV RLLRLGRVAR KLDHYLEYGA AVLVLLVCVF GLVAHWLACI WYSIGDYEVI DEVTNTIQID SWLYQLALSI GTPY RYNTS AGIWEGGPSK DSLYVSSLYF TMTSLTTIGF GNIAPTTDVE KMFSVAMMMV GSLLYATIFG NVTTIFQQMY ANTNR YHEM LNNVRDFLKL YQVPKGLSER VMDYIVSTWS MSKGIDTEKV LSICPKDMRA DICVHLNRKV FNEHPAFRLA SDGCLR ALA VEFQTIHCAP GDLIYHAGES VDALCFVVSG SLEVIQDDEV VAILGKGDVF GDIFWKETTL AHACANVRAL TYCDLHI IK REALLKVLDF YTAFANSFSR NLTLTCNLRK RIIFRKISDV KKEEEERLRQ KNEVTLSIPV DHPVRKLFQK FKQQKELR N QGSTQGDPER NQLQVESRSL QNGASITGTS VVTVSQITPI QTSLAYVKTS ESLKQNNRDA MELKPNGGAD QKCLKVNSP IRMKNGNGKG WLRLKNNMGA HEEKKEDWNN VTKAESMGLL SEDPKSSDSE NSVTKNPLRK TDSCDSGITK SDLRLDKAGE ARSPLEHSP IQADAKHPFY PIPEQALQTT LQEVKHELKE DIQLLSCRMT ALEKQVAEIL KILSEKSVPQ ASSPKSQMPL Q VPPQIPCQ DIFSVSRPES PESDKDEIHF UniProtKB: Voltage-gated delayed rectifier potassium channel KCNH5 |
-Macromolecule #2: POTASSIUM ION
| Macromolecule | Name: POTASSIUM ION / type: ligand / ID: 2 / Number of copies: 6 / Formula: K |
|---|---|
| Molecular weight | Theoretical: 39.098 Da |
-Macromolecule #3: water
| Macromolecule | Name: water / type: ligand / ID: 3 / Number of copies: 8 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 / Component - Concentration: 150.0 mM / Component - Formula: NaCl / Component - Name: sodium chloride |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Support film - Material: CARBON / Support film - topology: HOLEY |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.4 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 126938 |
| Initial angle assignment | Type: NOT APPLICABLE |
| Final angle assignment | Type: NOT APPLICABLE |
 Movie
Movie Controller
Controller















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)