+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Subtomogram average of 70S ribosome (11x11) | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | 70S / bacterial / RIBOSOME | |||||||||
| Biological species |  | |||||||||
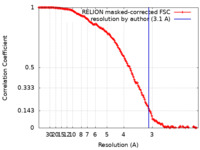
| Method | subtomogram averaging / cryo EM / Resolution: 3.1 Å | |||||||||
 Authors Authors | Eisenstein F / Danev R | |||||||||
| Funding support |  Japan, 2 items Japan, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Methods / Year: 2023 Journal: Nat Methods / Year: 2023Title: Parallel cryo electron tomography on in situ lamellae. Authors: Fabian Eisenstein / Haruaki Yanagisawa / Hiroka Kashihara / Masahide Kikkawa / Sachiko Tsukita / Radostin Danev /  Abstract: In situ cryo electron tomography of cryo focused ion beam milled samples has emerged in recent years as a powerful technique for structural studies of macromolecular complexes in their native ...In situ cryo electron tomography of cryo focused ion beam milled samples has emerged in recent years as a powerful technique for structural studies of macromolecular complexes in their native cellular environment. However, the possibilities for recording tomographic tilt series in a high-throughput manner are limited, in part by the lamella-shaped samples. Here we utilize a geometrical sample model and optical image shift to record tens of tilt series in parallel, thereby saving time and gaining access to sample areas conventionally used for tracking specimen movement. The parallel cryo electron tomography (PACE-tomo) method achieves a throughput faster than 5 min per tilt series and allows for the collection of sample areas that were previously unreachable, thus maximizing the amount of data from each lamella. Performance testing with ribosomes in vitro and in situ on state-of-the-art and general-purpose microscopes demonstrated the high throughput and quality of PACE-tomo. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_33834.map.gz emd_33834.map.gz | 142.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-33834-v30.xml emd-33834-v30.xml emd-33834.xml emd-33834.xml | 15.9 KB 15.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_33834_fsc.xml emd_33834_fsc.xml | 14.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_33834.png emd_33834.png | 128.5 KB | ||
| Masks |  emd_33834_msk_1.map emd_33834_msk_1.map | 244.1 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-33834.cif.gz emd-33834.cif.gz | 4.3 KB | ||
| Others |  emd_33834_half_map_1.map.gz emd_33834_half_map_1.map.gz emd_33834_half_map_2.map.gz emd_33834_half_map_2.map.gz | 119.7 MB 119.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-33834 http://ftp.pdbj.org/pub/emdb/structures/EMD-33834 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33834 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33834 | HTTPS FTP |
-Validation report
| Summary document |  emd_33834_validation.pdf.gz emd_33834_validation.pdf.gz | 1.1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_33834_full_validation.pdf.gz emd_33834_full_validation.pdf.gz | 1.1 MB | Display | |
| Data in XML |  emd_33834_validation.xml.gz emd_33834_validation.xml.gz | 22.3 KB | Display | |
| Data in CIF |  emd_33834_validation.cif.gz emd_33834_validation.cif.gz | 29 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33834 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33834 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33834 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33834 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_33834.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_33834.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.07 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_33834_msk_1.map emd_33834_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_33834_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_33834_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : 70S ribosome
| Entire | Name: 70S ribosome |
|---|---|
| Components |
|
-Supramolecule #1: 70S ribosome
| Supramolecule | Name: 70S ribosome / type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.7 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| |||||||||||||||
| Grid | Model: UltrAuFoil R1.2/1.3 / Mesh: 300 / Support film - Material: GOLD / Support film - topology: HOLEY | |||||||||||||||
| Vitrification | Cryogen name: ETHANE-PROPANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Detector mode: COUNTING / Number grids imaged: 1 / Average exposure time: 0.25 sec. / Average electron dose: 4.2 e/Å2 Details: 121 tilt series were recorded in parallel using beam image shift |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 3.0 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)