[English] 日本語
 Yorodumi
Yorodumi- EMDB-33825: SARS-CoV-2 spike in complex with neutralizing antibody NIV-10 (st... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | SARS-CoV-2 spike in complex with neutralizing antibody NIV-10 (state 2) | |||||||||||||||||||||||||||||||||
 Map data Map data | ||||||||||||||||||||||||||||||||||
 Sample Sample |
| |||||||||||||||||||||||||||||||||
 Keywords Keywords | Complex / VIRAL PROTEIN / VIRAL PROTEIN-IMMUNE SYSTEM complex | |||||||||||||||||||||||||||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | |||||||||||||||||||||||||||||||||
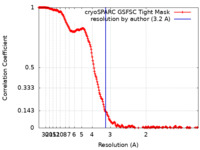
| Method | single particle reconstruction / cryo EM / Resolution: 3.2 Å | |||||||||||||||||||||||||||||||||
 Authors Authors | Moriyama S / Anraku Y / Muranishi S / Adachi Y / Kuroda D / Higuchi Y / Kotaki R / Tonouchi K / Yumoto K / Suzuki T ...Moriyama S / Anraku Y / Muranishi S / Adachi Y / Kuroda D / Higuchi Y / Kotaki R / Tonouchi K / Yumoto K / Suzuki T / Kita S / Fukuhara H / Kuroda Y / Yamamoto T / Onodera T / Fukushi S / Maeda K / Nakamura-Uchiyama F / Hashiguchi T / Hoshino A / Maenaka K / Takahashi Y | |||||||||||||||||||||||||||||||||
| Funding support |  Japan, 10 items Japan, 10 items
| |||||||||||||||||||||||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Structural delineation and computational design of SARS-CoV-2-neutralizing antibodies against Omicron subvariants. Authors: Saya Moriyama / Yuki Anraku / Shunta Taminishi / Yu Adachi / Daisuke Kuroda / Shunsuke Kita / Yusuke Higuchi / Yuhei Kirita / Ryutaro Kotaki / Keisuke Tonouchi / Kohei Yumoto / Tateki Suzuki ...Authors: Saya Moriyama / Yuki Anraku / Shunta Taminishi / Yu Adachi / Daisuke Kuroda / Shunsuke Kita / Yusuke Higuchi / Yuhei Kirita / Ryutaro Kotaki / Keisuke Tonouchi / Kohei Yumoto / Tateki Suzuki / Taiyou Someya / Hideo Fukuhara / Yudai Kuroda / Tsukasa Yamamoto / Taishi Onodera / Shuetsu Fukushi / Ken Maeda / Fukumi Nakamura-Uchiyama / Takao Hashiguchi / Atsushi Hoshino / Katsumi Maenaka / Yoshimasa Takahashi /  Abstract: SARS-CoV-2 Omicron subvariants have evolved to evade receptor-binding site (RBS) antibodies that exist in diverse individuals as public antibody clones. We rationally selected RBS antibodies ...SARS-CoV-2 Omicron subvariants have evolved to evade receptor-binding site (RBS) antibodies that exist in diverse individuals as public antibody clones. We rationally selected RBS antibodies resilient to mutations in emerging Omicron subvariants. Y489 was identified as a site of virus vulnerability and a common footprint of broadly neutralizing antibodies against the subvariants. Multiple Y489-binding antibodies were encoded by public clonotypes and additionally recognized F486, potentially accounting for the emergence of Omicron subvariants harboring the F486V mutation. However, a subclass of antibodies broadly neutralized BA.4/BA.5 variants via hydrophobic binding sites of rare clonotypes along with high mutation-resilience under escape mutation screening. A computationally designed antibody based on one of the Y489-binding antibodies, NIV-10/FD03, was able to bind XBB with any 486 mutation and neutralized XBB.1.5. The structural basis for the mutation-resilience of this Y489-binding antibody group may provide important insights into the design of therapeutics resistant to viral escape. | |||||||||||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_33825.map.gz emd_33825.map.gz | 362.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-33825-v30.xml emd-33825-v30.xml emd-33825.xml emd-33825.xml | 24.9 KB 24.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_33825_fsc.xml emd_33825_fsc.xml | 19.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_33825.png emd_33825.png | 68.4 KB | ||
| Masks |  emd_33825_msk_1.map emd_33825_msk_1.map | 729 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-33825.cif.gz emd-33825.cif.gz | 6.9 KB | ||
| Others |  emd_33825_half_map_1.map.gz emd_33825_half_map_1.map.gz emd_33825_half_map_2.map.gz emd_33825_half_map_2.map.gz | 676.1 MB 676.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-33825 http://ftp.pdbj.org/pub/emdb/structures/EMD-33825 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33825 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33825 | HTTPS FTP |
-Validation report
| Summary document |  emd_33825_validation.pdf.gz emd_33825_validation.pdf.gz | 1.2 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_33825_full_validation.pdf.gz emd_33825_full_validation.pdf.gz | 1.2 MB | Display | |
| Data in XML |  emd_33825_validation.xml.gz emd_33825_validation.xml.gz | 29 KB | Display | |
| Data in CIF |  emd_33825_validation.cif.gz emd_33825_validation.cif.gz | 37.8 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33825 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33825 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33825 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33825 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_33825.map.gz / Format: CCP4 / Size: 729 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_33825.map.gz / Format: CCP4 / Size: 729 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.67 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_33825_msk_1.map emd_33825_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_33825_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_33825_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : SARS-COV-2 spike glycoprotein in complex with NIV-10
| Entire | Name: SARS-COV-2 spike glycoprotein in complex with NIV-10 |
|---|---|
| Components |
|
-Supramolecule #1: SARS-COV-2 spike glycoprotein in complex with NIV-10
| Supramolecule | Name: SARS-COV-2 spike glycoprotein in complex with NIV-10 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Molecular weight | Theoretical: 570 KDa |
-Supramolecule #2: SARS-CoV-2 spike glycoprotein
| Supramolecule | Name: SARS-CoV-2 spike glycoprotein / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 420 KDa |
-Supramolecule #3: NIV-10 Fab
| Supramolecule | Name: NIV-10 Fab / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2-#3 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 50 KDa |
-Macromolecule #1: SARS-CoV-2 spike glycoprotein
| Macromolecule | Name: SARS-CoV-2 spike glycoprotein / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
| Sequence | String: SSQCVNLTTR TQLPPAYTNS FTRGVYYPDK VFRSSVLHST QDLFLPFFSN VTWFHAIHVS GTNGTKRFDN PVLPFNDGVY FASTEKSNII RGWIFGTTLD SKTQSLLIVN NATNVVIKVC EFQFCNDPFL GVYYHKNNKS WMESEFRVYS SANNCTFEYV SQPFLMDLEG ...String: SSQCVNLTTR TQLPPAYTNS FTRGVYYPDK VFRSSVLHST QDLFLPFFSN VTWFHAIHVS GTNGTKRFDN PVLPFNDGVY FASTEKSNII RGWIFGTTLD SKTQSLLIVN NATNVVIKVC EFQFCNDPFL GVYYHKNNKS WMESEFRVYS SANNCTFEYV SQPFLMDLEG KQGNFKNLRE FVFKNIDGYF KIYSKHTPIN LVRDLPQGFS ALEPLVDLPI GINITRFQTL LALHRSYLTP GDSSSGWTAG AAAYYVGYLQ PRTFLLKYNE NGTITDAVDC ALDPLSETKC TLKSFTVEKG IYQTSNFRVQ PTESIVRFPN ITNLCPFGEV FNATRFASVY AWNRKRISNC VADYSVLYNS ASFSTFKCYG VSPTKLNDLC FTNVYADSFV IRGDEVRQIA PGQTGKIADY NYKLPDDFTG CVIAWNSNNL DSKVGGNYNY LYRLFRKSNL KPFERDISTE IYQAGSTPCN GVEGFNCYFP LQSYGFQPTN GVGYQPYRVV VLSFELLHAP ATVCGPKKST NLVKNKCVNF NFNGLTGTGV LTESNKKFLP FQQFGRDIAD TTDAVRDPQT LEILDITPCS FGGVSVITPG TNTSNQVAVL YQDVNCTEVP VAIHADQLTP TWRVYSTGSN VFQTRAGCLI GAEHVNNSYE CDIPIGAGIC ASYQTQTQTN SPGSAGSVAS QSIIAYTMSL GAENSVAYSN NSIAIPTNFT ISVTTEILPV SMTKTSVDCT MYICGDSTEC SNLLLQYGSF CTQLNRALTG IAVEQDKNTQ EVFAQVKQIY KTPPIKDFGG FNFSQILPDP SKPSKRSPIE DLLFNKVTLA DAGFIKQYGD CLGDIAARDL ICAQKFNGLT VLPPLLTDEM IAQYTSALLA GTITSGWTFG AGPALQIPFP MQMAYRFNGI GVTQNVLYEN QKLIANQFNS AIGKIQDSLS STPSALGKLQ DVVNQNAQAL NTLVKQLSSN FGAISSVLND ILSRLDPPEA EVQIDRLITG RLQSLQTYVT QQLIRAAEIR ASANLAATKM SECVLGQSKR VDFCGKGYHL MSFPQSAPHG VVFLHVTYVP AQEKNFTTAP AICHDGKAHF PREGVFVSNG THWFVTQRNF YEPQIITTDN TFVSGNCDVV IGIVNNTVYD PLQPELDSFK EELDKYFKNH TSPDVDLGDI SGINASVVNI QKEIDRLNEV AKNLNESLID LQELGKYEQY I |
-Macromolecule #2: NIV-10 Fab light chain
| Macromolecule | Name: NIV-10 Fab light chain / type: protein_or_peptide / ID: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QSALTQPASV SGSPGQSITI SCTGTSSDVG GYNFVSWYRQ YPGKAPQLMI YDVSRRPSGD SDRFSGSKSG NTASLTISGL QAEDEAEYHC SSYTGRSPYV VFGGGTKVTV LRTVAAPSVF IFPPSDEQLK SGTASVVCLL NNFYPREAKV QWKVDNALQS GNSQESVTEQ ...String: QSALTQPASV SGSPGQSITI SCTGTSSDVG GYNFVSWYRQ YPGKAPQLMI YDVSRRPSGD SDRFSGSKSG NTASLTISGL QAEDEAEYHC SSYTGRSPYV VFGGGTKVTV LRTVAAPSVF IFPPSDEQLK SGTASVVCLL NNFYPREAKV QWKVDNALQS GNSQESVTEQ DSKDSTYSLS STLTLSKADY EKHKVYACEV THQGLSSPVT KSFNRGEC |
-Macromolecule #3: NIV-10 Fab heavy chain
| Macromolecule | Name: NIV-10 Fab heavy chain / type: protein_or_peptide / ID: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: EVQLVESGGG LVQPGGSLRL SCAASGFTFY SYWMTWVRQA PGKGLEWVAN IKHDESESHY VDSVRGRFTI SRDNAKNSVY LQMKGLRAED TAVYYCARDG GYNILTAYYH APSYWGQGTL VTVSSASTKG PSVFPLAPSS KSTSGGTAAL GCLVKDYFPE PVTVSWNSGA ...String: EVQLVESGGG LVQPGGSLRL SCAASGFTFY SYWMTWVRQA PGKGLEWVAN IKHDESESHY VDSVRGRFTI SRDNAKNSVY LQMKGLRAED TAVYYCARDG GYNILTAYYH APSYWGQGTL VTVSSASTKG PSVFPLAPSS KSTSGGTAAL GCLVKDYFPE PVTVSWNSGA LTSGVHTFPA VLQSSGLYSL SSVVTVPSSS LGTQTYICNV NHKPSNTKVD KKVEPKS |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.6 mg/mL |
|---|---|
| Buffer | pH: 7.4 |
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. / Pretreatment - Atmosphere: AIR |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 291 K / Instrument: FEI VITROBOT MARK IV / Details: blotting time 5 s and blotting force 5.. |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Digitization - Dimensions - Width: 5760 pixel / Digitization - Dimensions - Height: 4092 pixel / Number real images: 3110 / Average exposure time: 1.5 sec. / Average electron dose: 57.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 130000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller




















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)