+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of 1:1 PAPP-A.STC2 complex(half map) | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Hydrolase / METAL BINDING PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationregulation of hormone biosynthetic process / pappalysin-1 / response to follicle-stimulating hormone / regulation of store-operated calcium entry / response to vitamin D / negative regulation of multicellular organism growth / detection of maltose stimulus / response to dexamethasone / maltose transport complex / maltose binding ...regulation of hormone biosynthetic process / pappalysin-1 / response to follicle-stimulating hormone / regulation of store-operated calcium entry / response to vitamin D / negative regulation of multicellular organism growth / detection of maltose stimulus / response to dexamethasone / maltose transport complex / maltose binding / carbohydrate transport / maltose transport / maltodextrin transmembrane transport / decidualization / carbohydrate transmembrane transporter activity / ATP-binding cassette (ABC) transporter complex, substrate-binding subunit-containing / endoplasmic reticulum unfolded protein response / embryo implantation / ATP-binding cassette (ABC) transporter complex / cell chemotaxis / female pregnancy / Post-translational protein phosphorylation / protein catabolic process / hormone activity / metalloendopeptidase activity / response to peptide hormone / intracellular calcium ion homeostasis / Regulation of Insulin-like Growth Factor (IGF) transport and uptake by Insulin-like Growth Factor Binding Proteins (IGFBPs) / metallopeptidase activity / outer membrane-bounded periplasmic space / cellular response to hypoxia / response to oxidative stress / periplasmic space / cell surface receptor signaling pathway / endoplasmic reticulum lumen / negative regulation of gene expression / DNA damage response / heme binding / perinuclear region of cytoplasm / Golgi apparatus / enzyme binding / endoplasmic reticulum / protein homodimerization activity / proteolysis / extracellular space / zinc ion binding / extracellular region / membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
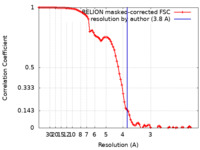
| Method | single particle reconstruction / cryo EM / Resolution: 3.8 Å | |||||||||
 Authors Authors | Zhong QH / Chu HL / Wang GP / Zhang C / Wei Y / Qiao J / Hang J | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Cell Discov / Year: 2022 Journal: Cell Discov / Year: 2022Title: Structural insights into the covalent regulation of PAPP-A activity by proMBP and STC2. Authors: Qihang Zhong / Honglei Chu / Guopeng Wang / Cheng Zhang / Rong Li / Fusheng Guo / Xinlu Meng / Xiaoguang Lei / Youli Zhou / Ruobing Ren / Lin Tao / Ningning Li / Ning Gao / Yuan Wei / Jie Qiao / Jing Hang /  Abstract: Originally discovered in the circulation of pregnant women as a protein secreted by placental trophoblasts, the metalloprotease pregnancy-associated plasma protein A (PAPP-A) is also widely expressed ...Originally discovered in the circulation of pregnant women as a protein secreted by placental trophoblasts, the metalloprotease pregnancy-associated plasma protein A (PAPP-A) is also widely expressed by many other tissues. It cleaves insulin-like growth factor-binding proteins (IGFBPs) to increase the bioavailability of IGFs and plays essential roles in multiple growth-promoting processes. While the vast majority of the circulatory PAPP-A in pregnancy is proteolytically inactive due to covalent inhibition by proform of eosinophil major basic protein (proMBP), the activity of PAPP-A can also be covalently inhibited by another less characterized modulator, stanniocalcin-2 (STC2). However, the structural basis of PAPP-A proteolysis and the mechanistic differences between these two modulators are poorly understood. Here we present two cryo-EM structures of endogenous purified PAPP-A in complex with either proMBP or STC2. Both modulators form 2:2 heterotetramer with PAPP-A and establish extensive interactions with multiple domains of PAPP-A that are distal to the catalytic cleft. This exosite-binding property results in a steric hindrance to prevent the binding and cleavage of IGFBPs, while the IGFBP linker region-derived peptides harboring the cleavage sites are no longer sensitive to the modulator treatment. Functional investigation into proMBP-mediated PAPP-A regulation in selective intrauterine growth restriction (sIUGR) pregnancy elucidates that PAPP-A and proMBP collaboratively regulate extravillous trophoblast invasion and the consequent fetal growth. Collectively, our work reveals a novel covalent exosite-competitive inhibition mechanism of PAPP-A and its regulatory effect on placental function. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_33622.map.gz emd_33622.map.gz | 5.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-33622-v30.xml emd-33622-v30.xml emd-33622.xml emd-33622.xml | 18 KB 18 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_33622_fsc.xml emd_33622_fsc.xml | 9.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_33622.png emd_33622.png | 48.2 KB | ||
| Filedesc metadata |  emd-33622.cif.gz emd-33622.cif.gz | 7.3 KB | ||
| Others |  emd_33622_half_map_1.map.gz emd_33622_half_map_1.map.gz emd_33622_half_map_2.map.gz emd_33622_half_map_2.map.gz | 49.5 MB 49.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-33622 http://ftp.pdbj.org/pub/emdb/structures/EMD-33622 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33622 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33622 | HTTPS FTP |
-Validation report
| Summary document |  emd_33622_validation.pdf.gz emd_33622_validation.pdf.gz | 664.1 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_33622_full_validation.pdf.gz emd_33622_full_validation.pdf.gz | 663.7 KB | Display | |
| Data in XML |  emd_33622_validation.xml.gz emd_33622_validation.xml.gz | 15.8 KB | Display | |
| Data in CIF |  emd_33622_validation.cif.gz emd_33622_validation.cif.gz | 20.6 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33622 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33622 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33622 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33622 | HTTPS FTP |
-Related structure data
| Related structure data |  7y5qMC  7y5nC  8hggC  8hghC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_33622.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_33622.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.055 Å | ||||||||||||||||||||||||||||||||||||
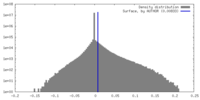
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_33622_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
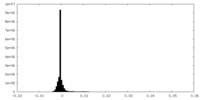
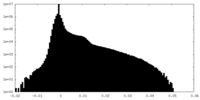
| Density Histograms |
-Half map: #1
| File | emd_33622_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
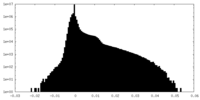
| Density Histograms |
- Sample components
Sample components
-Entire : Structure of 1:1 PAPP-A/STC2 complex(half map)
| Entire | Name: Structure of 1:1 PAPP-A/STC2 complex(half map) |
|---|---|
| Components |
|
-Supramolecule #1: Structure of 1:1 PAPP-A/STC2 complex(half map)
| Supramolecule | Name: Structure of 1:1 PAPP-A/STC2 complex(half map) / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Maltose/maltodextrin-binding periplasmic protein,Pappalysin-1
| Macromolecule | Name: Maltose/maltodextrin-binding periplasmic protein,Pappalysin-1 type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO / EC number: pappalysin-1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 216.398344 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: AASHHHHHHH HHHSGKIEEG KLVIWINGDK GYNGLAEVGK KFEKDTGIKV TVEHPDKLEE KFPQVAATGD GPDIIFWAHD RFGGYAQSG LLAEITPDKA FQDKLYPFTW DAVRYNGKLI AYPIAVEALS LIYNKDLLPN PPKTWEEIPA LDKELKAKGK S ALMFNLQE ...String: AASHHHHHHH HHHSGKIEEG KLVIWINGDK GYNGLAEVGK KFEKDTGIKV TVEHPDKLEE KFPQVAATGD GPDIIFWAHD RFGGYAQSG LLAEITPDKA FQDKLYPFTW DAVRYNGKLI AYPIAVEALS LIYNKDLLPN PPKTWEEIPA LDKELKAKGK S ALMFNLQE PYFTWPLIAA DGGYAFKYEN GKYDIKDVGV DNAGAKAGLT FLVDLIKNKH MNADTDYSIA EAAFNKGETA MT INGPWAW SNIDTSKVNY GVTVLPTFKG QPSKPFVGVL SAGINAASPN KELAKEFLEN YLLTDEGLEA VNKDKPLGAV ALK SYEEEL AKDPRIAATM ENAQKGEIMP NIPQMSAFWY AVRTAVINAA SGRQTVDEAL KDAQTDYDIP TTENLYFQGE FREA RGATE EPSPPSRALY FSGRGEQLRL RADLELPRDA FTLQVWLRAE GGQRSPAVIT GLYDKCSYIS RDRGWVVGIH TISDQ DNKD PRYFFSLKTD RARQVTTINA HRSYLPGQWV YLAATYDGQF MKLYVNGAQV ATSGEQVGGI FSPLTQKCKV LMLGGS ALN HNYRGYIEHF SLWKVARTQR EILSDMETHG AHTALPQLLL QENWDNVKHA WSPMKDGSSP KVEFSNAHGF LLDTSLE PP LCGQTLCDNT EVIASYNQLS SFRQPKVVRY RVVNLYEDDH KNPTVTREQV DFQHHQLAEA FKQYNISWEL DVLEVSNS S LRRRLILANC DISKIGDENC DPECNHTLTG HDGGDCRHLR HPAFVKKQHN GVCDMDCNYE RFNFDGGECC DPEITNVTQ TCFDPDSPHR AYLDVNELKN ILKLDGSTHL NIFFAKSSEE ELAGVATWPW DKEALMHLGG IVLNPSFYGM PGHTHTMIHE IGHSLGLYH VFRGISEIQS CSDPCMETEP SFETGDLCND TNPAPKHKSC GDPGPGNDTC GFHSFFNTPY NNFMSYADDD C TDSFTPNQ VARMHCYLDL VYQGWQPSRK PAPVALAPQV LGHTTDSVTL EWFPPIDGHF FERELGSACH LCLEGRILVQ YA SNASSPM PCSPSGHWSP REAEGHPDVE QPCKSSVRTW SPNSAVNPHT VPPACPEPQG CYLELEFLYP LVPESLTIWV TFV STDWDS SGAVNDIKLL AVSGKNISLG PQNVFCDVPL TIRLWDVGEE VYGIQIYTLD EHLEIDAAML TSTADTPLCL QCKP LKYKV VRDPPLQMDV ASILHLNRKF VDMDLNLGSV YQYWVITISG TEESEPSPAV TYIHGSGYCG DGIIQKDQGE QCDDM NKIN GDGCSLFCRQ EVSFNCIDEP SRCYFHDGDG VCEEFEQKTS IKDCGVYTPQ GFLDQWASNA SVSHQDQQCP GWVIIG QPA ASQVCRTKVI DLSEGISQHA WYPCTISYPY SQLAQTTFWL RAYFSQPMVA AAVIVHLVTD GTYYGDQKQE TISVQLL DT KDQSHDLGLH VLSCRNNPLI IPVVHDLSQP FYHSQAVRVS FSSPLVAISG VALRSFDNFD PVTLSSCQRG ETYSPAEQ S CVHFACEKTD CPELAVENAS LNCSSSDRYH GAQCTVSCRT GYVLQIRRDD ELIKSQTGPS VTVTCTEGKW NKQVACEPV DCSIPDHHQV YAASFSCPEG TTFGSQCSFQ CRHPAQLKGN NSLLTCMEDG LWSFPEALCE LMCLAPPPVP NADLQTARCR ENKHKVGSF CKYKCKPGYH VPGSSRKSKK RAFKTQCTQD GSWQEGACVP VTCDPPPPKF HGLYQCTNGF QFNSECRIKC E DSDASQGL GSNVIHCRKD GTWNGSFHVC QEMQGQCSVP NELNSNLKLQ CPDGYAIGSE CATSCLDHNS ESIILPMNVT VR DIPHWLN PTRVERVVCT AGLKWYPHPA LIHCVKGCEP FMGDNYCDAI NNRAFCNYDG GDCCTSTVKT KKVTPFPMSC DLQ GDCACR DPQAQEHSRK DLRGYSHG UniProtKB: Maltose/maltodextrin-binding periplasmic protein, Pappalysin-1 |
-Macromolecule #2: Stanniocalcin-2
| Macromolecule | Name: Stanniocalcin-2 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 33.298688 KDa |
| Sequence | String: MCAERLGQFM TLALVLATFD PARGTDATNP PEGPQDRSSQ QKGRLSLQNT AEIQHCLVNA GDVGCGVFEC FENNSCEIRG LHGICMTFL HNAGKFDAQG KSFIKDALKC KAHALRHRFG CISRKCPAIR EMVSQLQREC YLKHDLCAAA QENTRVIVEM I HFKDLLLH ...String: MCAERLGQFM TLALVLATFD PARGTDATNP PEGPQDRSSQ QKGRLSLQNT AEIQHCLVNA GDVGCGVFEC FENNSCEIRG LHGICMTFL HNAGKFDAQG KSFIKDALKC KAHALRHRFG CISRKCPAIR EMVSQLQREC YLKHDLCAAA QENTRVIVEM I HFKDLLLH EPYVDLVNLL LTCGEEVKEA ITHSVQVQCE QNWGSLCSIL SFCTSAIQKP PTAPPERQPQ VDRTKLSRAH HG EAGHHLP EPSSRETGRG AKGERGSKSH PNAHARGRVG GLGAQGPSGS SEWEDEQSEY SDIRR UniProtKB: Stanniocalcin-2 |
-Macromolecule #3: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 3 / Number of copies: 1 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Detector mode: COUNTING / Average electron dose: 59.8 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.2 µm / Nominal defocus min: 0.7000000000000001 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)