+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | structure of vp51 in white spot syndrome virus capsid | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Complex / C14 / capsid / disc / VIRAL PROTEIN | |||||||||
| Function / homology | viral envelope / DNA binding / Capsid protein Function and homology information Function and homology information | |||||||||
| Biological species |  White spot syndrome virus White spot syndrome virus | |||||||||
| Method | single particle reconstruction / cryo EM / negative staining / Resolution: 3.73 Å | |||||||||
 Authors Authors | Shan H / Liu MD / Shen QT | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: structure of vp51 in white spot syndrome virus capsid Authors: Hong S / Shen QT | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_33168.map.gz emd_33168.map.gz | 408 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-33168-v30.xml emd-33168-v30.xml emd-33168.xml emd-33168.xml | 12.9 KB 12.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_33168.png emd_33168.png | 143.9 KB | ||
| Masks |  emd_33168_msk_1.map emd_33168_msk_1.map | 824 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-33168.cif.gz emd-33168.cif.gz | 5 KB | ||
| Others |  emd_33168_half_map_1.map.gz emd_33168_half_map_1.map.gz emd_33168_half_map_2.map.gz emd_33168_half_map_2.map.gz | 764.6 MB 764.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-33168 http://ftp.pdbj.org/pub/emdb/structures/EMD-33168 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33168 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33168 | HTTPS FTP |
-Validation report
| Summary document |  emd_33168_validation.pdf.gz emd_33168_validation.pdf.gz | 1.1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_33168_full_validation.pdf.gz emd_33168_full_validation.pdf.gz | 1.1 MB | Display | |
| Data in XML |  emd_33168_validation.xml.gz emd_33168_validation.xml.gz | 20.7 KB | Display | |
| Data in CIF |  emd_33168_validation.cif.gz emd_33168_validation.cif.gz | 24.8 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33168 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33168 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33168 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33168 | HTTPS FTP |
-Related structure data
| Related structure data |  7xf2MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_33168.map.gz / Format: CCP4 / Size: 824 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_33168.map.gz / Format: CCP4 / Size: 824 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 1.34 Å | ||||||||||||||||||||
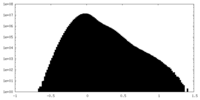
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_33168_msk_1.map emd_33168_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
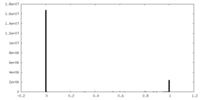
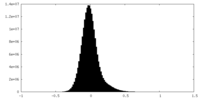
| Density Histograms |
-Half map: #2
| File | emd_33168_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
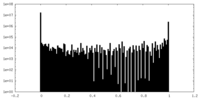
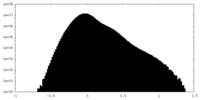
| Density Histograms |
-Half map: #1
| File | emd_33168_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
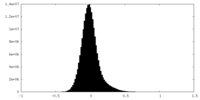
| Density Histograms |
- Sample components
Sample components
-Entire : white spot syndrome virus
| Entire | Name:  white spot syndrome virus white spot syndrome virus |
|---|---|
| Components |
|
-Supramolecule #1: white spot syndrome virus
| Supramolecule | Name: white spot syndrome virus / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  White spot syndrome virus White spot syndrome virus |
-Macromolecule #1: Capsid protein
| Macromolecule | Name: Capsid protein / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  White spot syndrome virus White spot syndrome virus |
| Molecular weight | Theoretical: 51.965145 KDa |
| Sequence | String: MSASLILDEY LKKTASAVLD VADSFEKIKG EIQSPEEAAA LSVALYGAPP KPSASAVASI ITGERTSLND KYLSDNVLLK MSVARVGQE NNRKRADQAA DEIRTIMEDI TGSLSGAYRQ YSPLEEENKV HIGIMNNKTP SIVCGYYTMD TSISSEPLSL T DFQNPTVI ...String: MSASLILDEY LKKTASAVLD VADSFEKIKG EIQSPEEAAA LSVALYGAPP KPSASAVASI ITGERTSLND KYLSDNVLLK MSVARVGQE NNRKRADQAA DEIRTIMEDI TGSLSGAYRQ YSPLEEENKV HIGIMNNKTP SIVCGYYTMD TSISSEPLSL T DFQNPTVI ANVTKRMESI FSKVDSARST RFDAFVNGVA NNMDIKSSID WANMVENVIK LPDSTPNPCS VDTIVSRDAS VV KTAVNDI YASVGKSYCR PATQLTFMSE IEKLRKAAVV CFEALMSDTR ERAFVEFLFY VSFKEDASNT NSKLFVQNKL SSM SGNPRQ PIKLVRRSAE ETLFGLCFMF KVMPPEFMNC IFNFPTIPHS TQYHGLYGTC LTPLLRKYGS SFEKSWAHFE EILS ERANA VKKFGVNDTR IDCLDAVANL TGPVYVLILD LVRTLSAQRS CSTKFLREIK ENYLLWNRFV S UniProtKB: Capsid protein |
-Experimental details
-Structure determination
| Method | negative staining, cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Staining | Type: NEGATIVE / Material: uranyl acetate |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 1.25 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 1.8 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: OTHER |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.73 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 153916 |
| Initial angle assignment | Type: PROJECTION MATCHING |
| Final angle assignment | Type: PROJECTION MATCHING |
 Movie
Movie Controller
Controller




 Z
Z Y
Y X
X