[English] 日本語
 Yorodumi
Yorodumi- EMDB-33113: Photobacterium phosphoreum fatty acid reductase complex LuxC-LuxE -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Photobacterium phosphoreum fatty acid reductase complex LuxC-LuxE | |||||||||
 Map data Map data | EM map of Photobacterium phosphoreum fatty acid reductase complex LuxC-LuxE | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | fatty acid reductase / acyl-protein synthetase / acyl-CoA reductase / bacterial bioluminescenc / LUMINESCENT PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationlong-chain acyl-protein thioester reductase / long-chain fatty acid--protein ligase activity / long-chain-fatty-acyl-CoA reductase activity / acyl-CoA dehydrogenase activity / bioluminescence Similarity search - Function | |||||||||
| Biological species |  Photobacterium phosphoreum (bacteria) Photobacterium phosphoreum (bacteria) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.79 Å | |||||||||
 Authors Authors | Tian Q / Huo Y / Wang L | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: J Biol Chem / Year: 2022 Journal: J Biol Chem / Year: 2022Title: Cryo-EM structure of the fatty acid reductase LuxC-LuxE complex provides insights into bacterial bioluminescence. Authors: Qingwei Tian / Jingting Wu / Haifeng Xu / Zhangli Hu / Yangao Huo / Liyan Wang /  Abstract: The discovery of reduced flavin mononucleotide and fatty aldehydes as essential factors of light emission facilitated study of bacterial luminescence. Although the molecular mechanisms underlying ...The discovery of reduced flavin mononucleotide and fatty aldehydes as essential factors of light emission facilitated study of bacterial luminescence. Although the molecular mechanisms underlying bacterial luminescence have been studied for more than 60 years, the structure of the bacterial fatty acid reductase complex remains unclear. Here, we report the cryo-EM structure of the Photobacterium phosphoreum fatty acid reductase complex LuxC-LuxE to a resolution of 2.79 Å. We show that the active site Lys238/Arg355 pair of LuxE is >30 Å from the active site Cys296 of LuxC, implying that catalysis relies on a large conformational change. Furthermore, mutagenesis and biochemical experiments support that the L-shaped cleft inside LuxC plays an important role in substrate binding and reaction. We obtained a series of mutants with significantly improved activity as measured by in vitro bioluminescence assays and demonstrated that the double mutant W111A/F483K displayed the highest activity (370% of the WT). Our results indicated that the activity of LuxC significantly affects the bacterial bioluminescence reaction. Finally, we expressed this mutated lux operon in Escherichia coli but observed that the in vivo concentrations of ATP and NADPH limited the enzyme activity; thus, we conclude that the luminous intensity mainly depends on the level of metabolic energy. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_33113.map.gz emd_33113.map.gz | 140.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-33113-v30.xml emd-33113-v30.xml emd-33113.xml emd-33113.xml | 15.7 KB 15.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_33113.png emd_33113.png | 30.3 KB | ||
| Filedesc metadata |  emd-33113.cif.gz emd-33113.cif.gz | 5.7 KB | ||
| Others |  emd_33113_half_map_1.map.gz emd_33113_half_map_1.map.gz emd_33113_half_map_2.map.gz emd_33113_half_map_2.map.gz | 118.4 MB 118.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-33113 http://ftp.pdbj.org/pub/emdb/structures/EMD-33113 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33113 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33113 | HTTPS FTP |
-Validation report
| Summary document |  emd_33113_validation.pdf.gz emd_33113_validation.pdf.gz | 986 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_33113_full_validation.pdf.gz emd_33113_full_validation.pdf.gz | 985.6 KB | Display | |
| Data in XML |  emd_33113_validation.xml.gz emd_33113_validation.xml.gz | 14.1 KB | Display | |
| Data in CIF |  emd_33113_validation.cif.gz emd_33113_validation.cif.gz | 16.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33113 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33113 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33113 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33113 | HTTPS FTP |
-Related structure data
| Related structure data |  7xc6MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_33113.map.gz / Format: CCP4 / Size: 149.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_33113.map.gz / Format: CCP4 / Size: 149.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | EM map of Photobacterium phosphoreum fatty acid reductase complex LuxC-LuxE | ||||||||||||||||||||
| Voxel size | X=Y=Z: 0.833 Å | ||||||||||||||||||||
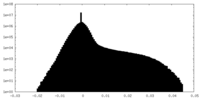
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: EM 2 half map of Photobacterium phosphoreum fatty...
| File | emd_33113_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | EM 2 half map of Photobacterium phosphoreum fatty acid reductase complex LuxC-LuxE | ||||||||||||
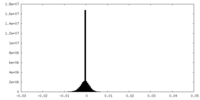
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: EM half map of Photobacterium phosphoreum fatty acid...
| File | emd_33113_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | EM half map of Photobacterium phosphoreum fatty acid reductase complex LuxC-LuxE | ||||||||||||
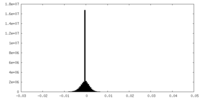
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : fatty acid reductase complex
| Entire | Name: fatty acid reductase complex |
|---|---|
| Components |
|
-Supramolecule #1: fatty acid reductase complex
| Supramolecule | Name: fatty acid reductase complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Photobacterium phosphoreum (bacteria) Photobacterium phosphoreum (bacteria) |
-Macromolecule #1: LuxE
| Macromolecule | Name: LuxE / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Photobacterium phosphoreum (bacteria) Photobacterium phosphoreum (bacteria) |
| Molecular weight | Theoretical: 42.934445 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MTITLDICEK DIIVSTEIDD IIFTSSPLDI TYDEQERIKH KLILESFRYH YNNNEDYKSF CNTQGVDENI SSLDDIPVFP TSMFKYAKI CTADESNIEN WFTSSGTSGV KSHIARDRVS IERLLGSVNY GMKYLGSFHE NQLELVNMGP DRFNAKNVWF K YVMSLVEL ...String: MTITLDICEK DIIVSTEIDD IIFTSSPLDI TYDEQERIKH KLILESFRYH YNNNEDYKSF CNTQGVDENI SSLDDIPVFP TSMFKYAKI CTADESNIEN WFTSSGTSGV KSHIARDRVS IERLLGSVNY GMKYLGSFHE NQLELVNMGP DRFNAKNVWF K YVMSLVEL LYPTTFTVNN DEIDFELTIK SLKEIYNKGK GICLIGPPYF IYLLCQYMKE NDIEFNAGNR IFIITGGGWK TK QKQALNR QDFNQLLMET FHLAHESQIR DTFNQVELNT CFFEDNRQRK HVPPWVYARA LDPVTLKPVE DGQEGLISYM DAS STSYPT FIVTDDIGII HTIKAPDPHQ GTTIDIVRRL NTREQKGCSL SMSSGLK UniProtKB: LuxE |
-Macromolecule #2: Long-chain acyl-protein thioester reductase
| Macromolecule | Name: Long-chain acyl-protein thioester reductase / type: protein_or_peptide / ID: 2 / Number of copies: 4 / Enantiomer: LEVO / EC number: long-chain acyl-protein thioester reductase |
|---|---|
| Source (natural) | Organism:  Photobacterium phosphoreum (bacteria) Photobacterium phosphoreum (bacteria) |
| Molecular weight | Theoretical: 54.067164 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: IKKIPMIIGG AERDTSEHEY RELTLNSYKV SIPIINQDDV EAIKSQSVEN NLNINQIVNF LYTVGQKWKS ENYSRRLTYI RDLVRFLGY SPEMAKLEAN WISMILSSKS ALYDIVETEL GSRHIVDEWL PQGDCYVKAM PKGKSVHLLA GNVPLSGVTS I IRAILTKN ...String: IKKIPMIIGG AERDTSEHEY RELTLNSYKV SIPIINQDDV EAIKSQSVEN NLNINQIVNF LYTVGQKWKS ENYSRRLTYI RDLVRFLGY SPEMAKLEAN WISMILSSKS ALYDIVETEL GSRHIVDEWL PQGDCYVKAM PKGKSVHLLA GNVPLSGVTS I IRAILTKN ECIIKTSSAD PFTAIALASS FIDTDEHHPI SRSMSVMYWS HNEDIAIPQQ IMNCADVVVS WGGYDAIKWA TE HTPVNVD ILKFGPKKSI AIVDNPVDIT ASAIGVAHDI CFYDQQACFS TQDIYYIGDN IDAFFDELVE QLNLYMDILP KGD QTFDEK ASFSLIEKEC QFAKYKVEKG DNQSWLLVKS PLGSFGNQPL ARSAYIHHVS DISEITPYIE NRITQTVTVT PWES SFKYR DVLASHGAER IVESGMNNIF RVGGAHDGMR PLQRLVKYIS HERPYTYSTK DVAVKIEQTR YLEEDKFLVF VP UniProtKB: Long-chain acyl-protein thioester reductase |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 40 sec. / Pretreatment - Atmosphere: AIR |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 2.79 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 229033 |
| Initial angle assignment | Type: ANGULAR RECONSTITUTION |
| Final angle assignment | Type: ANGULAR RECONSTITUTION |
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
|---|---|
| Output model |  PDB-7xc6: |
 Movie
Movie Controller
Controller





 Z
Z Y
Y X
X