[English] 日本語
 Yorodumi
Yorodumi- EMDB-33045: SARS-CoV-2 spike glycoprotein in complex with neutralizing antibo... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | SARS-CoV-2 spike glycoprotein in complex with neutralizing antibody UT28K | |||||||||||||||
 Map data Map data | ||||||||||||||||
 Sample Sample |
| |||||||||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | |||||||||||||||
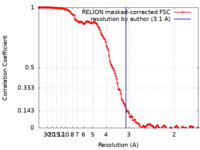
| Method | single particle reconstruction / cryo EM / Resolution: 3.1 Å | |||||||||||||||
 Authors Authors | Ozawa T / Tani H / Anraku Y / Kita S / Igarashi E / Saga Y / Inasaki N / Kawasuji H / Yamada H / Sasaki S ...Ozawa T / Tani H / Anraku Y / Kita S / Igarashi E / Saga Y / Inasaki N / Kawasuji H / Yamada H / Sasaki S / Someoka M / Sasaki J / Hayakawa Y / Yamamoto Y / Morinaga Y / Kurosawa N / Isobe M / Fukuhara H / Maenaka K / Hashiguchi T / Kishi H / Kitajima I / Saito S / Niimi H | |||||||||||||||
| Funding support |  Japan, 4 items Japan, 4 items
| |||||||||||||||
 Citation Citation |  Journal: MAbs / Year: 2022 Journal: MAbs / Year: 2022Title: Novel super-neutralizing antibody UT28K is capable of protecting against infection from a wide variety of SARS-CoV-2 variants. Authors: Tatsuhiko Ozawa / Hideki Tani / Yuki Anraku / Shunsuke Kita / Emiko Igarashi / Yumiko Saga / Noriko Inasaki / Hitoshi Kawasuji / Hiroshi Yamada / So-Ichiro Sasaki / Mayu Somekawa / Jiei ...Authors: Tatsuhiko Ozawa / Hideki Tani / Yuki Anraku / Shunsuke Kita / Emiko Igarashi / Yumiko Saga / Noriko Inasaki / Hitoshi Kawasuji / Hiroshi Yamada / So-Ichiro Sasaki / Mayu Somekawa / Jiei Sasaki / Yoshihiro Hayakawa / Yoshihiro Yamamoto / Yoshitomo Morinaga / Nobuyuki Kurosawa / Masaharu Isobe / Hideo Fukuhara / Katsumi Maenaka / Takao Hashiguchi / Hiroyuki Kishi / Isao Kitajima / Shigeru Saito / Hideki Niimi /  Abstract: Many potent neutralizing SARS-CoV-2 antibodies have been developed and used for therapies. However, the effectiveness of many antibodies has been reduced against recently emerging SARS-CoV-2 ...Many potent neutralizing SARS-CoV-2 antibodies have been developed and used for therapies. However, the effectiveness of many antibodies has been reduced against recently emerging SARS-CoV-2 variants, especially the Omicron variant. We identified a highly potent SARS-CoV-2 neutralizing antibody, UT28K, in COVID-19 convalescent individuals who recovered from a severe condition. UT28K showed efficacy in neutralizing SARS-CoV-2 in an assay and prophylactic treatment, and the reactivity to the Omicron strain was reduced. The structural analyses revealed that antibody UT28K Fab and SARS-CoV-2 RBD protein interactions were mainly chain-dominated antigen-antibody interactions. In addition, a mutation analysis suggested that the emergence of a UT28K neutralization-resistant SARS-CoV-2 variant was unlikely, as this variant would likely lose its competitive advantage over circulating SARS-CoV-2. Our data suggest that UT28K offers potent protection against SARS-CoV-2, including newly emerging variants. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_33045.map.gz emd_33045.map.gz | 170.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-33045-v30.xml emd-33045-v30.xml emd-33045.xml emd-33045.xml | 24.2 KB 24.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_33045_fsc.xml emd_33045_fsc.xml | 13.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_33045.png emd_33045.png | 99.8 KB | ||
| Masks |  emd_33045_msk_1.map emd_33045_msk_1.map | 216 MB |  Mask map Mask map | |
| Others |  emd_33045_half_map_1.map.gz emd_33045_half_map_1.map.gz emd_33045_half_map_2.map.gz emd_33045_half_map_2.map.gz | 171.2 MB 171.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-33045 http://ftp.pdbj.org/pub/emdb/structures/EMD-33045 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33045 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33045 | HTTPS FTP |
-Validation report
| Summary document |  emd_33045_validation.pdf.gz emd_33045_validation.pdf.gz | 819.3 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_33045_full_validation.pdf.gz emd_33045_full_validation.pdf.gz | 818.8 KB | Display | |
| Data in XML |  emd_33045_validation.xml.gz emd_33045_validation.xml.gz | 21 KB | Display | |
| Data in CIF |  emd_33045_validation.cif.gz emd_33045_validation.cif.gz | 27.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33045 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33045 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33045 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33045 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_33045.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_33045.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.85 Å | ||||||||||||||||||||||||||||||||||||
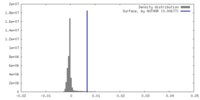
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_33045_msk_1.map emd_33045_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
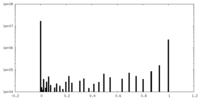
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_33045_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
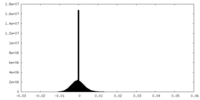
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_33045_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
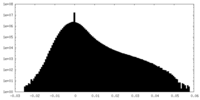
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : SARS-COV-2 spike glycoprotein in complex with UT28K
| Entire | Name: SARS-COV-2 spike glycoprotein in complex with UT28K |
|---|---|
| Components |
|
-Supramolecule #1: SARS-COV-2 spike glycoprotein in complex with UT28K
| Supramolecule | Name: SARS-COV-2 spike glycoprotein in complex with UT28K / type: complex / Chimera: Yes / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 50 KDa |
-Supramolecule #2: SARS-CoV-2 spike glycoprotein
| Supramolecule | Name: SARS-CoV-2 spike glycoprotein / type: complex / Chimera: Yes / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
-Supramolecule #3: UT28K Fab
| Supramolecule | Name: UT28K Fab / type: complex / Chimera: Yes / ID: 3 / Parent: 1 / Macromolecule list: #2-#3 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: SARS-CoV-2 spike glycoprotein
| Macromolecule | Name: SARS-CoV-2 spike glycoprotein / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
| Sequence | String: SSQCVNLTTR TQLPPAYTNS FTRGVYYPDK VFRSSVLHST QDLFLPFFSN VTWFHAIHVS GTNGTKRFDN PVLPFNDGVY FASTEKSNII RGWIFGTTLD SKTQSLLIVN NATNVVIKVC EFQFCNDPFL GVYYHKNNKS WMESEFRVYS SANNCTFEYV SQPFLMDLEG ...String: SSQCVNLTTR TQLPPAYTNS FTRGVYYPDK VFRSSVLHST QDLFLPFFSN VTWFHAIHVS GTNGTKRFDN PVLPFNDGVY FASTEKSNII RGWIFGTTLD SKTQSLLIVN NATNVVIKVC EFQFCNDPFL GVYYHKNNKS WMESEFRVYS SANNCTFEYV SQPFLMDLEG KQGNFKNLRE FVFKNIDGYF KIYSKHTPIN LVRDLPQGFS ALEPLVDLPI GINITRFQTL LALHRSYLTP GDSSSGWTAG AAAYYVGYLQ PRTFLLKYNE NGTITDAVDC ALDPLSETKC TLKSFTVEKG IYQTSNFRVQ PTESIVRFPN ITNLCPFGEV FNATRFASVY AWNRKRISNC VADYSVLYNS ASFSTFKCYG VSPTKLNDLC FTNVYADSFV IRGDEVRQIA PGQTGKIADY NYKLPDDFTG CVIAWNSNNL DSKVGGNYNY LYRLFRKSNL KPFERDISTE IYQAGSTPCN GVEGFNCYFP LQSYGFQPTN GVGYQPYRVV VLSFELLHAP ATVCGPKKST NLVKNKCVNF NFNGLTGTGV LTESNKKFLP FQQFGRDIAD TTDAVRDPQT LEILDITPCS FGGVSVITPG TNTSNQVAVL YQDVNCTEVP VAIHADQLTP TWRVYSTGSN VFQTRAGCLI GAEHVNNSYE CDIPIGAGIC ASYQTQTQTN SPGSAGSVAS QSIIAYTMSL GAENSVAYSN NSIAIPTNFT ISVTTEILPV SMTKTSVDCT MYICGDSTEC SNLLLQYGSF CTQLNRALTG IAVEQDKNTQ EVFAQVKQIY KTPPIKDFGG FNFSQILPDP SKPSKRSPIE DLLFNKVTLA DAGFIKQYGD CLGDIAARDL ICAQKFNGLT VLPPLLTDEM IAQYTSALLA GTITSGWTFG AGPALQIPFP MQMAYRFNGI GVTQNVLYEN QKLIANQFNS AIGKIQDSLS STPSALGKLQ DVVNQNAQAL NTLVKQLSSN FGAISSVLND ILSRLDPPEA EVQIDRLITG RLQSLQTYVT QQLIRAAEIR ASANLAATKM SECVLGQSKR VDFCGKGYHL MSFPQSAPHG VVFLHVTYVP AQEKNFTTAP AICHDGKAHF PREGVFVSNG THWFVTQRNF YEPQIITTDN TFVSGNCDVV IGIVNNTVYD PLQPELDSFK EELDKYFKNH TSPDVDLGDI SGINASVVNI QKEIDRLNEV AKNLNESLID LQELGKYEQY I |
-Macromolecule #2: UT28K Fab heavy chain
| Macromolecule | Name: UT28K Fab heavy chain / type: protein_or_peptide / ID: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MDWIWRILFL VGAATGAHSQ MQLVQSGPEV KKPGTSVKVS CKASGFTFSI SAVQWVRQAR GQRLEWIGWI VVGSGNTNYA QKFQERVTIT RDMSTSTAYM ELSSLRFEDT AVYYCAAPYC GGDCSDGFDI WGQGTMVTVS SASTKGPSVF PLAPSSKSTS GGTAALGCLV ...String: MDWIWRILFL VGAATGAHSQ MQLVQSGPEV KKPGTSVKVS CKASGFTFSI SAVQWVRQAR GQRLEWIGWI VVGSGNTNYA QKFQERVTIT RDMSTSTAYM ELSSLRFEDT AVYYCAAPYC GGDCSDGFDI WGQGTMVTVS SASTKGPSVF PLAPSSKSTS GGTAALGCLV KDYFPEPVTV SWNSGALTSG VHTFPAVLQS SGLYSLSSVV TVPSSSLGTQ TYICNVNHKP SNTKVDKKVE PKSGSSGHHH HHHHHHH |
-Macromolecule #3: UT28K Fab light chain
| Macromolecule | Name: UT28K Fab light chain / type: protein_or_peptide / ID: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: METPAQLLFL LLLWLPDTTG EIVLTQSPGT LSLSPGERAT LSCRASQSVS SSYLAWYQQK PGQAPRLLIY GASSRATGIP DRFSGSGSGT DFTLTISRLE PEDFAVYYCQ QYGNSPWTFG QGTKVEIKRT VAAPSVFIFP PSDEQLKSGT ASVVCLLNNF YPREAKVQWK ...String: METPAQLLFL LLLWLPDTTG EIVLTQSPGT LSLSPGERAT LSCRASQSVS SSYLAWYQQK PGQAPRLLIY GASSRATGIP DRFSGSGSGT DFTLTISRLE PEDFAVYYCQ QYGNSPWTFG QGTKVEIKRT VAAPSVFIFP PSDEQLKSGT ASVVCLLNNF YPREAKVQWK VDNALQSGNS QESVTEQDSK DSTYSLSSTL TLSKADYEKH KVYACEVTHQ GLSSPVTKSF NRGEC |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.1 mg/mL |
|---|---|
| Buffer | pH: 7.4 |
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Atmosphere: AIR |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 291 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Digitization - Dimensions - Width: 5760 pixel / Digitization - Dimensions - Height: 4092 pixel / Number real images: 2217 / Average exposure time: 1.5 sec. / Average electron dose: 53.14 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 105000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)