[English] 日本語
 Yorodumi
Yorodumi- EMDB-32725: Mouse TRPM8 in lipid nanodiscs in the presence of calcium, icilin... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Mouse TRPM8 in lipid nanodiscs in the presence of calcium, icilin and PI(4,5)P2 | |||||||||
 Map data Map data | nanodisc-ca-icilin-pip2 | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | TRPM8 / TRANSPORT PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationligand-gated calcium channel activity / TRP channels / thermoception / response to temperature stimulus / monoatomic ion channel activity / response to cold / calcium ion transmembrane transport / calcium channel activity / intracellular calcium ion homeostasis / calcium ion transport ...ligand-gated calcium channel activity / TRP channels / thermoception / response to temperature stimulus / monoatomic ion channel activity / response to cold / calcium ion transmembrane transport / calcium channel activity / intracellular calcium ion homeostasis / calcium ion transport / positive regulation of cold-induced thermogenesis / membrane raft / external side of plasma membrane / identical protein binding / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.04 Å | |||||||||
 Authors Authors | Zhao C / Xie Y | |||||||||
| Funding support |  China, 2 items China, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: Structures of a mammalian TRPM8 in closed state. Authors: Cheng Zhao / Yuan Xie / Lizhen Xu / Fan Ye / Ximing Xu / Wei Yang / Fan Yang / Jiangtao Guo /  Abstract: Transient receptor potential melastatin 8 (TRPM8) channel is a Ca-permeable non-selective cation channel that acts as the primary cold sensor in humans. TRPM8 is also activated by ligands such as ...Transient receptor potential melastatin 8 (TRPM8) channel is a Ca-permeable non-selective cation channel that acts as the primary cold sensor in humans. TRPM8 is also activated by ligands such as menthol, icilin, and phosphatidylinositol 4,5-bisphosphate (PIP), and desensitized by Ca. Here we have determined electron cryo-microscopy structures of mouse TRPM8 in the absence of ligand, and in the presence of Ca and icilin at 2.5-3.2 Å resolution. The ligand-free state TRPM8 structure represents the full-length structure of mammalian TRPM8 channels with a canonical S4-S5 linker and the clearly resolved selectivity filter and outer pore loop. TRPM8 has a short but wide selectivity filter which may account for its permeability to hydrated Ca. Ca and icilin bind in the cytosolic-facing cavity of the voltage-sensing-like domain of TRPM8 but induce little conformational change. All the ligand-bound TRPM8 structures adopt the same closed conformation as the ligand-free structure. This study reveals the overall architecture of mouse TRPM8 and the structural basis for its ligand recognition. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_32725.map.gz emd_32725.map.gz | 55 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-32725-v30.xml emd-32725-v30.xml emd-32725.xml emd-32725.xml | 11.3 KB 11.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_32725.png emd_32725.png | 96.4 KB | ||
| Filedesc metadata |  emd-32725.cif.gz emd-32725.cif.gz | 5.9 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-32725 http://ftp.pdbj.org/pub/emdb/structures/EMD-32725 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32725 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32725 | HTTPS FTP |
-Validation report
| Summary document |  emd_32725_validation.pdf.gz emd_32725_validation.pdf.gz | 573.1 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_32725_full_validation.pdf.gz emd_32725_full_validation.pdf.gz | 572.7 KB | Display | |
| Data in XML |  emd_32725_validation.xml.gz emd_32725_validation.xml.gz | 6.1 KB | Display | |
| Data in CIF |  emd_32725_validation.cif.gz emd_32725_validation.cif.gz | 7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32725 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32725 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32725 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32725 | HTTPS FTP |
-Related structure data
| Related structure data |  7wrfMC  7wraC  7wrbC  7wrcC  7wrdC  7wreC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_32725.map.gz / Format: CCP4 / Size: 59.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_32725.map.gz / Format: CCP4 / Size: 59.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | nanodisc-ca-icilin-pip2 | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.014 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : Mouse TRPM8 in lipid nanodiscs in the presence of calcium, icilin...
| Entire | Name: Mouse TRPM8 in lipid nanodiscs in the presence of calcium, icilin and PI(4,5)P2 |
|---|---|
| Components |
|
-Supramolecule #1: Mouse TRPM8 in lipid nanodiscs in the presence of calcium, icilin...
| Supramolecule | Name: Mouse TRPM8 in lipid nanodiscs in the presence of calcium, icilin and PI(4,5)P2 type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Transient receptor potential cation channel subfamily M member 8
| Macromolecule | Name: Transient receptor potential cation channel subfamily M member 8 type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 129.542227 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MSFEGARLSM RSRRNGTMGS TRTLYSSVSR STDVSYSDSD LVNFIQANFK KRECVFFTRD SKAMENICKC GYAQSQHIEG TQINQNEKW NYKKHTKEFP TDAFGDIQFE TLGKKGKYLR LSCDTDSETL YELLTQHWHL KTPNLVISVT GGAKNFALKP R MRKIFSRL ...String: MSFEGARLSM RSRRNGTMGS TRTLYSSVSR STDVSYSDSD LVNFIQANFK KRECVFFTRD SKAMENICKC GYAQSQHIEG TQINQNEKW NYKKHTKEFP TDAFGDIQFE TLGKKGKYLR LSCDTDSETL YELLTQHWHL KTPNLVISVT GGAKNFALKP R MRKIFSRL IYIAQSKGAW ILTGGTHYGL MKYIGEVVRD NTISRNSEEN IVAIGIAAWG MVSNRDTLIR SCDDEGHFSA QY IMDDFTR DPLYILDNNH THLLLVDNGC HGHPTVEAKL RNQLEKYISE RTSQDSNYGG KIPIVCFAQG GGRETLKAIN TSV KSKIPC VVVEGSGQIA DVIASLVEVE DVLTSSMVKE KLVRFLPRTV SRLPEEEIES WIKWLKEILE SSHLLTVIKM EEAG DEIVS NAISYALYKA FSTNEQDKDN WNGQLKLLLE WNQLDLASDE IFTNDRRWES ADLQEVMFTA LIKDRPKFVR LFLEN GLNL QKFLTNEVLT ELFSTHFSTL VYRNLQIAKN SYNDALLTFV WKLVANFRRS FWKEDRSSRE DLDVELHDAS LTTRHP LQA LFIWAILQNK KELSKVIWEQ TKGCTLAALG ASKLLKTLAK VKNDINAAGE SEELANEYET RAVELFTECY SNDEDLA EQ LLVYSCEAWG GSNCLELAVE ATDQHFIAQP GVQNFLSKQW YGEISRDTKN WKIILCLFII PLVGCGLVSF RKKPIDKH K KLLWYYVAFF TSPFVVFSWN VVFYIAFLLL FAYVLLMDFH SVPHTPELIL YALVFVLFCD EVRQWYMNGV NYFTDLWNV MDTLGLFYFI AGIVFRLHSS NKSSLYSGRV IFCLDYIIFT LRLIHIFTVS RNLGPKIIML QRMLIDVFFF LFLFAVWMVA FGVARQGIL RQNEQRWRWI FRSVIYEPYL AMFGQVPSDV DSTTYDFSHC TFSGNESKPL CVELDEHNLP RFPEWITIPL V CIYMLSTN ILLVNLLVAM FGYTVGIVQE NNDQVWKFQR YFLVQEYCNR LNIPFPFVVF AYFYMVVKKC FKCCCKEKNM ES NACCFRN EDNETLAWEG VMKENYLVKI NTKANDNSEE MRHRFRQLDS KLNDLKSLLK EIANNIKLEG GSSGGWSHPQ FEK UniProtKB: Transient receptor potential cation channel subfamily M member 8 |
-Macromolecule #2: CALCIUM ION
| Macromolecule | Name: CALCIUM ION / type: ligand / ID: 2 / Number of copies: 4 / Formula: CA |
|---|---|
| Molecular weight | Theoretical: 40.078 Da |
-Macromolecule #3: SODIUM ION
| Macromolecule | Name: SODIUM ION / type: ligand / ID: 3 / Number of copies: 1 |
|---|---|
| Molecular weight | Theoretical: 22.99 Da |
-Macromolecule #4: Icilin
| Macromolecule | Name: Icilin / type: ligand / ID: 4 / Number of copies: 4 / Formula: KX7 |
|---|---|
| Molecular weight | Theoretical: 311.292 Da |
| Chemical component information | 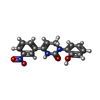 ChemComp-KX7: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 62.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: DARK FIELD / Nominal defocus max: 1.3 µm / Nominal defocus min: 1.1 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: OTHER |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.04 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 62791 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)




















