[English] 日本語
 Yorodumi
Yorodumi- EMDB-32630: Structure and dynamics of Odinarchaeota tubulin and the implicati... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure and dynamics of Odinarchaeota tubulin and the implications for eukaryotic microtubule evolution | ||||||||||||
 Map data Map data | Reconstruction of tubes of OdinTubulin sampled from Yellowstone Lower Culex Basin hot spring polymerized at 37 degrees Celsius | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | Eukaryotic tubulin homolog / STRUCTURAL PROTEIN | ||||||||||||
| Biological species |  Candidatus Odinarchaeota archaeon LCB_4 (archaea) Candidatus Odinarchaeota archaeon LCB_4 (archaea) | ||||||||||||
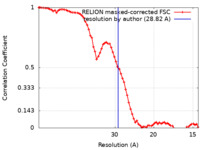
| Method | helical reconstruction / cryo EM / Resolution: 28.82 Å | ||||||||||||
 Authors Authors | Akil C / Ali S / Tran LT | ||||||||||||
| Funding support | 3 items
| ||||||||||||
 Citation Citation |  Journal: Sci Adv / Year: 2022 Journal: Sci Adv / Year: 2022Title: Structure and dynamics of Odinarchaeota tubulin and the implications for eukaryotic microtubule evolution. Authors: Caner Akıl / Samson Ali / Linh T Tran / Jérémie Gaillard / Wenfei Li / Kenichi Hayashida / Mika Hirose / Takayuki Kato / Atsunori Oshima / Kosuke Fujishima / Laurent Blanchoin / Akihiro ...Authors: Caner Akıl / Samson Ali / Linh T Tran / Jérémie Gaillard / Wenfei Li / Kenichi Hayashida / Mika Hirose / Takayuki Kato / Atsunori Oshima / Kosuke Fujishima / Laurent Blanchoin / Akihiro Narita / Robert C Robinson /     Abstract: Tubulins are critical for the internal organization of eukaryotic cells, and understanding their emergence is an important question in eukaryogenesis. Asgard archaea are the closest known prokaryotic ...Tubulins are critical for the internal organization of eukaryotic cells, and understanding their emergence is an important question in eukaryogenesis. Asgard archaea are the closest known prokaryotic relatives to eukaryotes. Here, we elucidated the apo and nucleotide-bound x-ray structures of an Asgard tubulin from hydrothermal living Odinarchaeota (OdinTubulin). The guanosine 5'-triphosphate (GTP)-bound structure resembles a microtubule protofilament, with GTP bound between subunits, coordinating the "+" end subunit through a network of water molecules and unexpectedly by two cations. A water molecule is located suitable for GTP hydrolysis. Time course crystallography and electron microscopy revealed conformational changes on GTP hydrolysis. OdinTubulin forms tubules at high temperatures, with short curved protofilaments coiling around the tubule circumference, more similar to FtsZ, rather than running parallel to its length, as in microtubules. Thus, OdinTubulin represents an evolutionary stage intermediate between prokaryotic FtsZ and eukaryotic microtubule-forming tubulins. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_32630.map.gz emd_32630.map.gz | 3.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-32630-v30.xml emd-32630-v30.xml emd-32630.xml emd-32630.xml | 14.9 KB 14.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_32630_fsc.xml emd_32630_fsc.xml | 7.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_32630.png emd_32630.png | 34 KB | ||
| Filedesc metadata |  emd-32630.cif.gz emd-32630.cif.gz | 5.8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-32630 http://ftp.pdbj.org/pub/emdb/structures/EMD-32630 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32630 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32630 | HTTPS FTP |
-Validation report
| Summary document |  emd_32630_validation.pdf.gz emd_32630_validation.pdf.gz | 379.7 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_32630_full_validation.pdf.gz emd_32630_full_validation.pdf.gz | 379.3 KB | Display | |
| Data in XML |  emd_32630_validation.xml.gz emd_32630_validation.xml.gz | 9.3 KB | Display | |
| Data in CIF |  emd_32630_validation.cif.gz emd_32630_validation.cif.gz | 12.1 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32630 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32630 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32630 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32630 | HTTPS FTP |
-Related structure data
| Related structure data |  7evbC  7evcC  7evdC  7eveC  7evgC  7evhC  7eviC  7evkC  7evlC  7f1aC  7f1bC C: citing same article ( |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_32630.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_32630.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Reconstruction of tubes of OdinTubulin sampled from Yellowstone Lower Culex Basin hot spring polymerized at 37 degrees Celsius | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 7.25 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : Tubulin homolog from Candidatus Odinarchaeota archaeon
| Entire | Name: Tubulin homolog from Candidatus Odinarchaeota archaeon |
|---|---|
| Components |
|
-Supramolecule #1: Tubulin homolog from Candidatus Odinarchaeota archaeon
| Supramolecule | Name: Tubulin homolog from Candidatus Odinarchaeota archaeon type: complex / ID: 1 / Parent: 0 / Macromolecule list: all Details: OdinTubulin was sampled from Candidatus Odinarchaeota archaeon LCB_4 MAG whose habitat is Yellowstone Lower Culex Basin hot spring. |
|---|---|
| Source (natural) | Organism:  Candidatus Odinarchaeota archaeon LCB_4 (archaea) Candidatus Odinarchaeota archaeon LCB_4 (archaea) |
| Molecular weight | Theoretical: 47.6 kDa/nm |
-Macromolecule #1: OdinTubulin
| Macromolecule | Name: OdinTubulin / type: other / ID: 1 Details: Candidatus Odinarchaeota archaeon LCB_4 (Odin) metagenome-assembled genome Classification: other |
|---|---|
| Source (natural) | Organism:  Candidatus Odinarchaeota archaeon LCB_4 (archaea) Candidatus Odinarchaeota archaeon LCB_4 (archaea) |
| Sequence | String: PMPGREILVL HVGQGGNQIG YNFWKTICEE HNIDIRSNQR KSVEEDKVDY KSVFLVEAPD GFHPRALFID LEPLAVEFLV KEMKLGSFFS EDLMVLSYSG AHNVWSIGYQ TGKKLIPVIL EKIRDTMPET LQGFLIIHTL GGGTGSGFGS LLTETLKKEF PGKGVLNFSV ...String: PMPGREILVL HVGQGGNQIG YNFWKTICEE HNIDIRSNQR KSVEEDKVDY KSVFLVEAPD GFHPRALFID LEPLAVEFLV KEMKLGSFFS EDLMVLSYSG AHNVWSIGYQ TGKKLIPVIL EKIRDTMPET LQGFLIIHTL GGGTGSGFGS LLTETLKKEF PGKGVLNFSV LPSEVNDVTL APYNTVLSLN HLSRFSDLVV LFDNTALIRI VKDQLNYPVI KQFSDLNFLI GRVMASITAS LRFPGPLNMD LMEMAHNLVA LPETKFIIPS VAPLTKEESE MSTELDLVER CFDPTHYMVN CSGQGKTISS VLMFRGNIAI ENAFSIMTDI KSNVAFAPGV HPDLGLKYGI CESAPVDFDK EVTLLSNNTI ISEVFNRVLE RFDSLFNRDW YTSHYVNAGT SKSNLKEARD NFDRIIKIYK EIEGSQ GENBANK: GENBANK: MDVT00000000.1 |
| Recombinant expression | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Concentration | 6.8 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 6.9 Component:
Details: 100 mM PIPES, pH 6.9, 0.5 mM EGTA, 0.5 mM MgSO4, 10% (v/v) glycerol and polymerized with 2 mM GTP | |||||||||||||||
| Grid | Model: Quantifoil R2/2 / Material: MOLYBDENUM / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 40 sec. / Pretreatment - Atmosphere: AIR / Details: High vacuum | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Chamber temperature: 298 K / Instrument: LEICA EM GP |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: COUNTING / Number grids imaged: 2 / Number real images: 1217 / Average exposure time: 2.42 sec. / Average electron dose: 40.05 e/Å2 Details: Images were recorded using a Falcon3 detector (FEI) at a pixel size of 1.45 angstrom per pixel and a frame rate of 60 frames/individual images |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.6 µm / Nominal defocus min: 2.0 µm / Nominal magnification: 47000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)