[English] 日本語
 Yorodumi
Yorodumi- EMDB-32516: SARS-CoV-2 spike protein in complex with three human neutralizing... -
+ Open data
Open data
- Basic information
Basic information
| Entry | 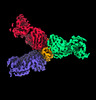 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
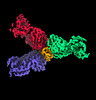
| Title | SARS-CoV-2 spike protein in complex with three human neutralizing antibodies | |||||||||
 Map data Map data | SARS-CoV-2 spike protein in complex with three human neutralizing antibodies | |||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationMaturation of spike protein / viral translation / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / suppression by virus of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion / entry receptor-mediated virion attachment to host cell ...Maturation of spike protein / viral translation / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / suppression by virus of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion / entry receptor-mediated virion attachment to host cell / host cell endoplasmic reticulum-Golgi intermediate compartment membrane / receptor-mediated endocytosis of virus by host cell / membrane fusion / Attachment and Entry / positive regulation of viral entry into host cell / receptor-mediated virion attachment to host cell / receptor ligand activity / host cell surface receptor binding / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / virion attachment to host cell / SARS-CoV-2 activates/modulates innate and adaptive immune responses / host cell plasma membrane / virion membrane / identical protein binding / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.42 Å | |||||||||
 Authors Authors | Zheng Q / Li S / Sun H / Zheng Z / Wang S | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Cell Rep / Year: 2022 Journal: Cell Rep / Year: 2022Title: Three SARS-CoV-2 antibodies provide broad and synergistic neutralization against variants of concern, including Omicron. Authors: Siling Wang / Hui Sun / Yali Zhang / Lunzhi Yuan / Yizhen Wang / Tianying Zhang / Shaojuan Wang / Jinlei Zhang / Hai Yu / Hualong Xiong / Zimin Tang / Liqin Liu / Yang Huang / Xiuting Chen / ...Authors: Siling Wang / Hui Sun / Yali Zhang / Lunzhi Yuan / Yizhen Wang / Tianying Zhang / Shaojuan Wang / Jinlei Zhang / Hai Yu / Hualong Xiong / Zimin Tang / Liqin Liu / Yang Huang / Xiuting Chen / Tingting Li / Dong Ying / Chang Liu / Zihao Chen / Quan Yuan / Jun Zhang / Tong Cheng / Shaowei Li / Yi Guan / Qingbing Zheng / Zizheng Zheng / Ningshao Xia /  Abstract: The rapidly spreading Omicron variant is highly resistant to vaccines, convalescent sera, and neutralizing antibodies (nAbs), highlighting the urgent need for potent therapeutic nAbs. Here, a panel ...The rapidly spreading Omicron variant is highly resistant to vaccines, convalescent sera, and neutralizing antibodies (nAbs), highlighting the urgent need for potent therapeutic nAbs. Here, a panel of human nAbs from severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) convalescent patients show diverse neutralization against Omicron, of which XMA01 and XMA04 maintain nanomolar affinities and excellent neutralization (half maximal inhibitory concentration [IC50]: ∼20 ng/mL). nAb XMA09 shows weak but unattenuated neutralization against all variants of concern (VOCs) as well as SARS-CoV. Structural analysis reveals that the above three antibodies could synergistically bind to the receptor-binding domains (RBDs) of both wild-type and Omicron spikes and defines the critical determinants for nAb-mediated broad neutralizations. Three nAbs confer synergistic neutralization against Omicron, resulting from the inter-antibody interaction between XMA04 and XMA01(or XMA09). Furthermore, the XMA01/XMA04 cocktail provides synergistic protection against Beta and Omicron variant infections in hamsters. In summary, our results provide insights for the rational design of antibody cocktail therapeutics or universal vaccines against Omicron. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_32516.map.gz emd_32516.map.gz | 230.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-32516-v30.xml emd-32516-v30.xml emd-32516.xml emd-32516.xml | 17.1 KB 17.1 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_32516.png emd_32516.png | 76.3 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-32516 http://ftp.pdbj.org/pub/emdb/structures/EMD-32516 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32516 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32516 | HTTPS FTP |
-Validation report
| Summary document |  emd_32516_validation.pdf.gz emd_32516_validation.pdf.gz | 412.3 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_32516_full_validation.pdf.gz emd_32516_full_validation.pdf.gz | 411.9 KB | Display | |
| Data in XML |  emd_32516_validation.xml.gz emd_32516_validation.xml.gz | 7.2 KB | Display | |
| Data in CIF |  emd_32516_validation.cif.gz emd_32516_validation.cif.gz | 8.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32516 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32516 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32516 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32516 | HTTPS FTP |
-Related structure data
| Related structure data |  7whzMC  7wi0C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_32516.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_32516.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | SARS-CoV-2 spike protein in complex with three human neutralizing antibodies | ||||||||||||||||||||
| Voxel size | X=Y=Z: 0.778 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : SARS-CoV-2 spike protein in complex with three human neutralizing...
| Entire | Name: SARS-CoV-2 spike protein in complex with three human neutralizing antibodies |
|---|---|
| Components |
|
-Supramolecule #1: SARS-CoV-2 spike protein in complex with three human neutralizing...
| Supramolecule | Name: SARS-CoV-2 spike protein in complex with three human neutralizing antibodies type: complex / Chimera: Yes / ID: 1 / Parent: 0 / Macromolecule list: #1-#7 |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Spike protein S1
| Macromolecule | Name: Spike protein S1 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 21.873496 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: TNLCPFGEVF NATRFASVYA WNRKRISNCV ADYSVLYNSA SFSTFKCYGV SPTKLNDLCF TNVYADSFVI RGDEVRQIAP GQTGKIADY NYKLPDDFTG CVIAWNSNNL DSKVGGNYNY LYRLFRKSNL KPFERDISTE IYQAGSTPCN GVEGFNCYFP L QSYGFQPT ...String: TNLCPFGEVF NATRFASVYA WNRKRISNCV ADYSVLYNSA SFSTFKCYGV SPTKLNDLCF TNVYADSFVI RGDEVRQIAP GQTGKIADY NYKLPDDFTG CVIAWNSNNL DSKVGGNYNY LYRLFRKSNL KPFERDISTE IYQAGSTPCN GVEGFNCYFP L QSYGFQPT NGVGYQPYRV VVLSFELLHA PATVCGP |
-Macromolecule #2: XMA04 heavy chain variable domain
| Macromolecule | Name: XMA04 heavy chain variable domain / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 13.517036 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: EVQLVESGGG VVQPGRSLRL SCAASGFTFR TYAMHWVRQA PGKGLEWVAV MWNDGSNKYY ADSVKGRFTI SRDNSKNTLY LEMNSLRAE DTAVYYCARE GVAAAGSSSD AFDIWGQGTM VTVSS |
-Macromolecule #3: XMA01 heavy chain variable domain
| Macromolecule | Name: XMA01 heavy chain variable domain / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 13.198615 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: EVQLVESGGG LVQPGGSLRL SCSASGFTFS SYAMHWVRQA PGKGLEYVSS ISSNGGSTYY ADSVKGRFTI SRDNSKNTLY LQMSSLRVE DAAVYYCVKH YQQQLLWGGP DVWGQGTTVT VSS |
-Macromolecule #4: IGL c4029_light_IGKV1-39_IGKJ2
| Macromolecule | Name: IGL c4029_light_IGKV1-39_IGKJ2 / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 11.616873 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: DIQMTQSPSS LSASVGDRVT ITCRASQSIS NFLNWYQQKP GKAPKLLIYA ASSLQSGVPS RFSGSGSGTD FTLTISSLQP EDFATYYCQ QSYSTLYSFG QGTKLEIK |
-Macromolecule #5: IG c934_light_IGKV1-5_IGKJ1
| Macromolecule | Name: IG c934_light_IGKV1-5_IGKJ1 / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 11.781073 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: DIQMTQSPVS LSASVGDRVT ITCRASQSIS TWLAWYQQKP GKAPKLLIYE TSSLESGVPS RFSGSGSGTE FTLTISSLQP DDFATYYCQ QFNSYPWTFG QGTKVEIK |
-Macromolecule #6: XMA09 heavy chain variable domain
| Macromolecule | Name: XMA09 heavy chain variable domain / type: protein_or_peptide / ID: 6 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 13.272664 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QVQLVQSGAE VKKPGSSVKV SCKASGGTFS SFAISWVRQA PGQGLEWMGG IIPIYGTANY AQKFQGRVTI TADESTSTAY MDLSSLRSE DTAVYYCARD GSSTYYPHNW FDPWGQGTLV TVSS |
-Macromolecule #7: IG c1437_light_IGLV1-40_IGLJ1
| Macromolecule | Name: IG c1437_light_IGLV1-40_IGLJ1 / type: protein_or_peptide / ID: 7 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 11.374391 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QSVLTQPPSV SGAPGQRVTI SCTGSSSNIG SGYDVHWYQQ LPGTAPKLLI YGNSNRPSGV PDRFSGSKSG TSASLAITGL QAEDEADYY CQSYDSSLSG VFGTGTKVTV L |
-Macromolecule #8: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 8 / Number of copies: 1 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F30 |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Tecnai F30 / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.42 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 721486 |
|---|---|
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller