+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of SaCas9-sgRNA-DNA ternary complex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationmaintenance of CRISPR repeat elements / defense response to virus / endonuclease activity / Hydrolases; Acting on ester bonds / DNA binding / RNA binding / metal ion binding Similarity search - Function | |||||||||
| Biological species |  | |||||||||
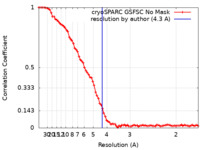
| Method | single particle reconstruction / cryo EM / Resolution: 4.3 Å | |||||||||
 Authors Authors | Du WH / Huang Q / Zhu HX / Xue DM / Zheng S | |||||||||
| Funding support |  China, 2 items China, 2 items
| |||||||||
 Citation Citation |  Journal: Int J Mol Sci / Year: 2023 Journal: Int J Mol Sci / Year: 2023Title: Full-Length Model of SaCas9-sgRNA-DNA Complex in Cleavage State. Authors: Wenhao Du / Haixia Zhu / Jiaqiang Qian / Dongmei Xue / Sen Zheng / Qiang Huang /  Abstract: Cas9 (SaCas9) is a widely used genome editing tool. Understanding its molecular mechanisms of DNA cleavage could effectively guide the engineering optimization of this system. Here, we determined ... Cas9 (SaCas9) is a widely used genome editing tool. Understanding its molecular mechanisms of DNA cleavage could effectively guide the engineering optimization of this system. Here, we determined the first cryo-electron microscopy structure of the SaCas9-sgRNA-DNA ternary complex. This structure reveals that the HNH nuclease domain is tightly bound to the cleavage site of the target DNA strand, and is in close contact with the WED and REC domains. Moreover, it captures the complete structure of the sgRNA, including the previously unresolved stem-loop 2. Based on this structure, we build a full-length model for the ternary complex in cleavage state. This model enables identification of the residues for the interactions between the HNH domain and the WED and REC domains. Moreover, we found that the stem-loop 2 of the sgRNA tightly binds to the PI and RuvC domains and may also regulate the position shift of the RuvC domain. Further mutagenesis and molecular dynamics simulations supported the idea that the interactions of the HNH domain with the WED and REC domains play an important role in the DNA cleavage. Thus, this study provides new mechanistic insights into the DNA cleavage of SaCas9 and is also useful for guiding the future engineering of SaCas9-mediated gene editing systems. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_32104.map.gz emd_32104.map.gz | 45.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-32104-v30.xml emd-32104-v30.xml emd-32104.xml emd-32104.xml | 17 KB 17 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_32104_fsc.xml emd_32104_fsc.xml | 8.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_32104.png emd_32104.png | 88.8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-32104 http://ftp.pdbj.org/pub/emdb/structures/EMD-32104 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32104 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32104 | HTTPS FTP |
-Validation report
| Summary document |  emd_32104_validation.pdf.gz emd_32104_validation.pdf.gz | 469.9 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_32104_full_validation.pdf.gz emd_32104_full_validation.pdf.gz | 469.5 KB | Display | |
| Data in XML |  emd_32104_validation.xml.gz emd_32104_validation.xml.gz | 10 KB | Display | |
| Data in CIF |  emd_32104_validation.cif.gz emd_32104_validation.cif.gz | 12.8 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32104 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32104 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32104 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32104 | HTTPS FTP |
-Related structure data
| Related structure data |  7vw3MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_32104.map.gz / Format: CCP4 / Size: 48.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_32104.map.gz / Format: CCP4 / Size: 48.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.85 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : SaCas9-sgRNA-target DNA ternary complex
| Entire | Name: SaCas9-sgRNA-target DNA ternary complex |
|---|---|
| Components |
|
-Supramolecule #1: SaCas9-sgRNA-target DNA ternary complex
| Supramolecule | Name: SaCas9-sgRNA-target DNA ternary complex / type: complex / ID: 1 / Chimera: Yes / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
-Supramolecule #2: SaCas9
| Supramolecule | Name: SaCas9 / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|
-Supramolecule #3: sgRNA-target DNA
| Supramolecule | Name: sgRNA-target DNA / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2-#4 |
|---|
-Macromolecule #1: SaCas9
| Macromolecule | Name: SaCas9 / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
| Sequence | String: MGSSHHHHHH SSGLVPRGSH MKRNYILGLA IGITSVGYGI IDYETRDVID AGVRLFKEAN VENNEGRRSK RGARRLKRRR RHRIQRVKKL LFDYNLLTDH SELSGINPYE ARVKGLSQKL SEEEFSAALL HLAKRRGVHN VNEVEEDTGN ELSTKEQISR NSKALEEKYV ...String: MGSSHHHHHH SSGLVPRGSH MKRNYILGLA IGITSVGYGI IDYETRDVID AGVRLFKEAN VENNEGRRSK RGARRLKRRR RHRIQRVKKL LFDYNLLTDH SELSGINPYE ARVKGLSQKL SEEEFSAALL HLAKRRGVHN VNEVEEDTGN ELSTKEQISR NSKALEEKYV AELQLERLKK DGEVRGSINR FKTSDYVKEA KQLLKVQKAY HQLDQSFIDT YIDLLETRRT YYEGPGEGSP FGWKDIKEWY EMLMGHCTYF PEELRSVKYA YNADLYNALN DLNNLVITRD ENEKLEYYEK FQIIENVFKQ KKKPTLKQIA KEILVNEEDI KGYRVTSTGK PEFTNLKVYH DIKDITARKE IIENAELLDQ IAKILTIYQS SEDIQEELTN LNSELTQEEI EQISNLKGYT GTHNLSLKAI NLILDELWHT NDNQIAIFNR LKLVPKKVDL SQQKEIPTTL VDDFILSPVV KRSFIQSIKV INAIIKKYGL PNDIIIELAR EKNSKDAQKM INEMQKRNRQ TNERIEEIIR TTGKENAKYL IEKIKLHDMQ EGKCLYSLEA IPLEDLLNNP FNYEVDHIIP RSVSFDNSFN NKVLVKQEEA SKKGNRTPFQ YLSSSDSKIS YETFKKHILN LAKGKGRISK TKKEYLLEER DINRFSVQKD FINRNLVDTR YATRGLMNLL RSYFRVNNLD VKVKSINGGF TSFLRRKWKF KKERNKGYKH HAEDALIIAN ADFIFKEWKK LDKAKKVMEN QMFEEKQAES MPEIETEQEY KEIFITPHQI KHIKDFKDYK YSHRVDKKPN RELINDTLYS TRKDDKGNTL IVNNLNGLYD KDNDKLKKLI NKSPEKLLMY HHDPQTYQKL KLIMEQYGDE KNPLYKYYEE TGNYLTKYSK KDNGPVIKKI KYYGNKLNAH LDITDDYPNS RNKVVKLSLK PYRFDVYLDN GVYKFVTVKN LDVIKKENYY EVNSKCYEEA KKLKKISNQA EFIASFYNND LIKINGELYR VIGVNNDLLN RIEVNMIDIT YREYLENMND KRPPRIIKTI ASKTQSIKKY STDILGNLYE VKSKKHPQII KKG |
-Macromolecule #2: single-guide RNA (sgRNA)
| Macromolecule | Name: single-guide RNA (sgRNA) / type: rna / ID: 2 |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Sequence | String: GGUACCGCUC CAGUCGUUCA UGGUUUUAGU ACUCUGGAAA CAGAAUCUAC UAAAACAAGG CAAAAUGCCG UGUUUAUCUC GUCAACUUGU UGGCGAGAUU UUUUU |
-Macromolecule #3: Target DNA strand(TS)
| Macromolecule | Name: Target DNA strand(TS) / type: dna / ID: 3 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Sequence | String: TCCAGAGTAC TAAAACATTC AACATGAACG ACTGGAGCGG TACTATAGTG AGTCGTATT |
-Macromolecule #4: Non-target DNA strand (NTS)
| Macromolecule | Name: Non-target DNA strand (NTS) / type: dna / ID: 4 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Sequence | String: AATACGACTC ACTATAGTAC CGCTCCAGTC GTTCATGTTG AATGTTTTAG TACTCTGGA |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.22 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
Details: 20mM Tris-Cl (pH 7.5), 100mM KCl, 5mM MgCl2, 1mM DTT | |||||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 40 sec. | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 289 K / Instrument: FEI VITROBOT MARK IV | |||||||||||||||
| Details | blot for 4 seconds before pluging |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Digitization - Dimensions - Width: 5760 pixel / Digitization - Dimensions - Height: 4092 pixel / Number real images: 10389 / Average electron dose: 38.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.6 µm / Nominal defocus min: 1.8 µm / Nominal magnification: 105000 |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)