+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | T.thermophilus 30S ribosome with KsgA, class K4 | ||||||||||||
 Map data Map data | B-factor sharpened map from the postprocess routine in Relion | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | 30S subunit / KsgA / rRNA methyltransferase / RIBOSOME | ||||||||||||
| Function / homology |  Function and homology information Function and homology information16S rRNA (adenine1518-N6/adenine1519-N6)-dimethyltransferase / 16S rRNA (adenine(1518)-N(6)/adenine(1519)-N(6))-dimethyltransferase activity / rRNA (adenine-N6,N6-)-dimethyltransferase activity / rRNA methylation / ribosomal small subunit biogenesis / ribosomal small subunit assembly / small ribosomal subunit / small ribosomal subunit rRNA binding / cytosolic small ribosomal subunit / tRNA binding ...16S rRNA (adenine1518-N6/adenine1519-N6)-dimethyltransferase / 16S rRNA (adenine(1518)-N(6)/adenine(1519)-N(6))-dimethyltransferase activity / rRNA (adenine-N6,N6-)-dimethyltransferase activity / rRNA methylation / ribosomal small subunit biogenesis / ribosomal small subunit assembly / small ribosomal subunit / small ribosomal subunit rRNA binding / cytosolic small ribosomal subunit / tRNA binding / rRNA binding / ribosome / structural constituent of ribosome / translation / ribonucleoprotein complex / mRNA binding / RNA binding / zinc ion binding / metal ion binding / cytosol / cytoplasm Similarity search - Function | ||||||||||||
| Biological species |   Thermus thermophilus HB8 (bacteria) / Thermus thermophilus HB8 (bacteria) /  | ||||||||||||
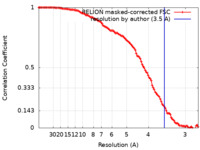
| Method | single particle reconstruction / cryo EM / Resolution: 3.5 Å | ||||||||||||
 Authors Authors | Raina R / Singh J | ||||||||||||
| Funding support |  India, 3 items India, 3 items
| ||||||||||||
 Citation Citation |  Journal: ACS Chem Biol / Year: 2022 Journal: ACS Chem Biol / Year: 2022Title: Decoding the Mechanism of Specific RNA Targeting by Ribosomal Methyltransferases. Authors: Juhi Singh / Rahul Raina / Kutti R Vinothkumar / Ruchi Anand /  Abstract: Methylation of specific nucleotides is integral for ribosomal biogenesis and also serves as a common mechanism to confer antibiotic resistance by pathogenic bacteria. Here, by determining the high- ...Methylation of specific nucleotides is integral for ribosomal biogenesis and also serves as a common mechanism to confer antibiotic resistance by pathogenic bacteria. Here, by determining the high-resolution structure of the 30S-KsgA complex by cryo-electron microscopy, a state was captured, where KsgA juxtaposes between helices h44 and h45 of the 30S ribosome, separating them, thereby enabling remodeling of the surrounded rRNA and allowing the cognate site to enter the methylation pocket. With the structure as a guide, several mutant versions of the ribosomes, where interacting bases in the catalytic helix h45 and surrounding helices h44, h24, and h27, were mutated and evaluated for their methylation efficiency revealing factors that direct the enzyme to its cognate site with high fidelity. The biochemical studies show that the three-dimensional environment of the ribosome enables the interaction of select loop regions in KsgA with the ribosome helices paramount to maintain selectivity. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_31658.map.gz emd_31658.map.gz | 117 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-31658-v30.xml emd-31658-v30.xml emd-31658.xml emd-31658.xml | 56.5 KB 56.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_31658_fsc.xml emd_31658_fsc.xml | 11.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_31658.png emd_31658.png | 43.3 KB | ||
| Filedesc metadata |  emd-31658.cif.gz emd-31658.cif.gz | 10.4 KB | ||
| Others |  emd_31658_additional_1.map.gz emd_31658_additional_1.map.gz emd_31658_additional_2.map.gz emd_31658_additional_2.map.gz emd_31658_additional_3.map.gz emd_31658_additional_3.map.gz emd_31658_additional_4.map.gz emd_31658_additional_4.map.gz emd_31658_additional_5.map.gz emd_31658_additional_5.map.gz emd_31658_half_map_1.map.gz emd_31658_half_map_1.map.gz emd_31658_half_map_2.map.gz emd_31658_half_map_2.map.gz | 5.4 MB 6.3 MB 8.1 MB 116.9 MB 13.2 MB 98.3 MB 98.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-31658 http://ftp.pdbj.org/pub/emdb/structures/EMD-31658 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-31658 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-31658 | HTTPS FTP |
-Validation report
| Summary document |  emd_31658_validation.pdf.gz emd_31658_validation.pdf.gz | 978.3 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_31658_full_validation.pdf.gz emd_31658_full_validation.pdf.gz | 977.8 KB | Display | |
| Data in XML |  emd_31658_validation.xml.gz emd_31658_validation.xml.gz | 18.8 KB | Display | |
| Data in CIF |  emd_31658_validation.cif.gz emd_31658_validation.cif.gz | 24.6 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-31658 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-31658 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-31658 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-31658 | HTTPS FTP |
-Related structure data
| Related structure data |  7v2oMC  7v2lC  7v2mC  7v2nC  7v2pC  7v2qC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_31658.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_31658.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | B-factor sharpened map from the postprocess routine in Relion | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.38 Å | ||||||||||||||||||||||||||||||||||||
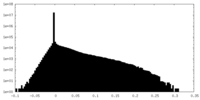
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: One of the maps from multibody refinement from...
| File | emd_31658_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | One of the maps from multibody refinement from Relion. This map was used in model building. | ||||||||||||
| Projections & Slices |
| ||||||||||||
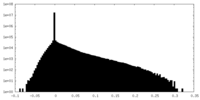
| Density Histograms |
-Additional map: One of the maps from multibody refinement from...
| File | emd_31658_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | One of the maps from multibody refinement from Relion. This map was used in model building. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: One of the maps from multibody refinement from...
| File | emd_31658_additional_3.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | One of the maps from multibody refinement from Relion. This map was used in model building. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Composite map of the multibody refinement from Phenix...
| File | emd_31658_additional_4.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Composite map of the multibody refinement from Phenix software. This map was used in model building. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: This map was calculated using LocSpiral. This map...
| File | emd_31658_additional_5.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | This map was calculated using LocSpiral. This map was used in model building. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: One of the half maps from 3D refine,...
| File | emd_31658_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | One of the half maps from 3D refine, used in the postprocess routine to obtain the primary map. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: One of the half maps from 3D refine,...
| File | emd_31658_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | One of the half maps from 3D refine, used in the postprocess routine to obtain the primary map. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
+Entire : 30S ribosomal with bound KsgA, a RNA methyltransferase
+Supramolecule #1: 30S ribosomal with bound KsgA, a RNA methyltransferase
+Supramolecule #2: 30S ribosomal subunit
+Supramolecule #3: KsgA
+Macromolecule #1: 16s ribosomal RNA
+Macromolecule #2: 30S ribosomal protein S2
+Macromolecule #3: 30S ribosomal protein S3
+Macromolecule #4: 30S ribosomal protein S4
+Macromolecule #5: 30S ribosomal protein S5
+Macromolecule #6: 30S ribosomal protein S6
+Macromolecule #7: 30S ribosomal protein S7
+Macromolecule #8: 30S ribosomal protein S8
+Macromolecule #9: 30S ribosomal protein S9
+Macromolecule #10: 30S ribosomal protein S10
+Macromolecule #11: 30S ribosomal protein S11
+Macromolecule #12: 30S ribosomal protein S12
+Macromolecule #13: 30S ribosomal protein S13
+Macromolecule #14: 30S ribosomal protein S14 type Z
+Macromolecule #15: 30S ribosomal protein S15
+Macromolecule #16: 30S ribosomal protein S16
+Macromolecule #17: 30S ribosomal protein S17
+Macromolecule #18: 30S ribosomal protein S18
+Macromolecule #19: 30S ribosomal protein S19
+Macromolecule #20: 30S ribosomal protein S20
+Macromolecule #21: Ribosomal RNA small subunit methyltransferase A
+Macromolecule #22: 30S ribosomal protein Thx
+Macromolecule #23: ZINC ION
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 Component:
| ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Atmosphere: AIR | ||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV Details: After application of the complex, 15 seconds wait time was given before blotting.. | ||||||||||
| Details | ~2 micromolar concentration of ribosome+KsgA was used during freezing |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: INTEGRATING / Digitization - Dimensions - Width: 4096 pixel / Digitization - Dimensions - Height: 4096 pixel / Number grids imaged: 1 / Average exposure time: 2.0 sec. / Average electron dose: 43.0 e/Å2 Details: Total number of frames is 30. Each frame had 1.43 e/A2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Calibrated magnification: 101449 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.5 µm / Nominal defocus min: 1.8 µm / Nominal magnification: 59000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Source name: PDB / Chain - Initial model type: experimental model |
|---|---|
| Details | Phenix was used in real space for initial refinement. The final refinements was performed with REFMAC within ccpem |
| Refinement | Space: RECIPROCAL / Protocol: OTHER / Overall B value: 109.3 |
| Output model |  PDB-7v2o: |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)