+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
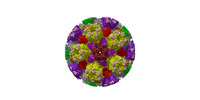
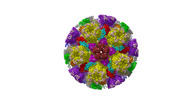
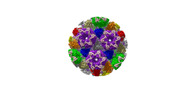
| Title | Cryo-EM structure of PRV A-capid | |||||||||
 Map data Map data | ||||||||||
 Sample Sample | Cryo-EM structure of PRV A-capsid != Suid alphaherpesvirus 1 Cryo-EM structure of PRV A-capsid
| |||||||||
| Function / homology |  Function and homology information Function and homology informationT=16 icosahedral viral capsid / viral capsid assembly / viral process / viral capsid / host cell nucleus / structural molecule activity / DNA binding Similarity search - Function | |||||||||
| Biological species |   Suid alphaherpesvirus 1 Suid alphaherpesvirus 1 | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.53 Å | |||||||||
 Authors Authors | Zheng Q / Li S / Zha Z / Sun H | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: Structures of pseudorabies virus capsids. Authors: Guosong Wang / Zhenghui Zha / Pengfei Huang / Hui Sun / Yang Huang / Maozhou He / Tian Chen / Lina Lin / Zhenqin Chen / Zhibo Kong / Yuqiong Que / Tingting Li / Ying Gu / Hai Yu / Jun Zhang ...Authors: Guosong Wang / Zhenghui Zha / Pengfei Huang / Hui Sun / Yang Huang / Maozhou He / Tian Chen / Lina Lin / Zhenqin Chen / Zhibo Kong / Yuqiong Que / Tingting Li / Ying Gu / Hai Yu / Jun Zhang / Qingbing Zheng / Yixin Chen / Shaowei Li / Ningshao Xia /  Abstract: Pseudorabies virus (PRV) is a major etiological agent of swine infectious diseases and is responsible for significant economic losses in the swine industry. Recent data points to human viral ...Pseudorabies virus (PRV) is a major etiological agent of swine infectious diseases and is responsible for significant economic losses in the swine industry. Recent data points to human viral encephalitis caused by PRV infection, suggesting that PRV may be able to overcome the species barrier to infect humans. To date, there is no available therapeutic for PRV infection. Here, we report the near-atomic structures of the PRV A-capsid and C-capsid, and illustrate the interaction that occurs between these subunits. We show that the C-capsid portal complex is decorated with capsid-associated tegument complexes. The PRV capsid structure is highly reminiscent of other α-herpesviruses, with some additional structural features of β- and γ-herpesviruses. These results illustrate the structure of the PRV capsid and elucidate the underlying assembly mechanism at the molecular level. This knowledge may be useful for the development of oncolytic agents or specific therapeutics against this arm of the herpesvirus family. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_31612.map.gz emd_31612.map.gz | 7.2 GB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-31612-v30.xml emd-31612-v30.xml emd-31612.xml emd-31612.xml | 13.6 KB 13.6 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_31612.png emd_31612.png | 34.2 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-31612 http://ftp.pdbj.org/pub/emdb/structures/EMD-31612 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-31612 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-31612 | HTTPS FTP |
-Validation report
| Summary document |  emd_31612_validation.pdf.gz emd_31612_validation.pdf.gz | 675.4 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_31612_full_validation.pdf.gz emd_31612_full_validation.pdf.gz | 675 KB | Display | |
| Data in XML |  emd_31612_validation.xml.gz emd_31612_validation.xml.gz | 13.7 KB | Display | |
| Data in CIF |  emd_31612_validation.cif.gz emd_31612_validation.cif.gz | 15.8 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-31612 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-31612 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-31612 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-31612 | HTTPS FTP |
-Related structure data
| Related structure data |  7fj3MC  7fj1C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_31612.map.gz / Format: CCP4 / Size: 7.8 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_31612.map.gz / Format: CCP4 / Size: 7.8 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 1.117 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : Cryo-EM structure of PRV A-capsid
| Entire | Name: Cryo-EM structure of PRV A-capsid |
|---|---|
| Components |
|
-Supramolecule #1: Suid alphaherpesvirus 1
| Supramolecule | Name: Suid alphaherpesvirus 1 / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all / NCBI-ID: 10345 / Sci species name: Suid alphaherpesvirus 1 / Virus type: VIRION / Virus isolate: OTHER / Virus enveloped: Yes / Virus empty: No |
|---|
-Macromolecule #1: Major capsid protein
| Macromolecule | Name: Major capsid protein / type: protein_or_peptide / ID: 1 / Number of copies: 16 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Suid alphaherpesvirus 1 Suid alphaherpesvirus 1 |
| Molecular weight | Theoretical: 146.101984 KDa |
| Sequence | String: MERPAILPSG QILSNIEVHS HRALFDIFKR FRSDDNNLYG AEFDALLGTY CSTLSLVRFL ELGLSVACVC TKFPELSYVA EGTIQFEVQ QPMIARDGPH PADQPVHNYM IKRLDRRSLN AAFSIAVEAL GLISGENLDG THISSAMRLR AIQQLARNVQ A VLDSFERG ...String: MERPAILPSG QILSNIEVHS HRALFDIFKR FRSDDNNLYG AEFDALLGTY CSTLSLVRFL ELGLSVACVC TKFPELSYVA EGTIQFEVQ QPMIARDGPH PADQPVHNYM IKRLDRRSLN AAFSIAVEAL GLISGENLDG THISSAMRLR AIQQLARNVQ A VLDSFERG TADQMLRVLM EKAPPLSLLA PFTLYEGRLA DRVACAALVS ELKRRVRDDT FFLTKHERNK DAVLDRLSDL VN CTAPSVA VARMTHADTQ GRPVDGVLVT TAGVRQRLLH HVLTLADTHA DVPVTYGEMV IANTNLVTAL VMGKAVSNMD DVA RYLLGG EPAPDDGKPV GSARVRADLV VVGDRLVFLE ALEKRVYQAT QVPYPLVGNL DVTFVMPLGV FKPAADRYAR HAGS FAPTP GLPDPRTHPP RAVHFFNKDG VPCHVTFEHA MGTLCHPSFL DVDATLAALR QEPAEVQCAF GAYVADARPD ALVGL MQRF LEEWPGMMPV RPRWAAPAAA DQLLAPGNAD LRLELHPAFD FFVAPEVDVP GPFAVPQVMG QVRAMPRIIN GNIPLA LCP VDFRDARGFE LSVDRHRLAP ATVAAVRGAF RDANYPMVFY IIEAVIHGSE RTFCALARLV AQCIQSYWRN THNAAFV NN FYMVMYINTY LGNGELPEDC AAVYKDLLEH VHALRRLIGE FTLPGDPLGN QPQEELNHAL ADATLLPPLI WDCDPILY R DGLAERLPEL RVNGAHFQHI LWVEMAQVNF RNVGGGLVHN RPVRNENQPL HPHHDAEWSV LSKIYYYAVV PAFSRGNCC TMGVRYDRVY QLVQTMVVPE TDEEVGTDDP RHPLHPRNLV PNSLNVLFHN ACVAVDADAM LILQETVTNM AERTTPLLAS VAPDAGMAT VATRDMRTHD GSLHHGLLMM AYQPNDATLL EGAFFYPAPV NALFACADHL GAMRDVGAEV RAAAQHVPCV P HFLGANYY ATVRQPVAQH AAQSRADENT LSYALMAGYF KMSPVAFTHQ LRRQLHPGFA LTVVRQDRFA TENVLFAEKA SE SYFMGQM QVARTESGGG LHLQLTQPRA NVDLGVGFTA AYAAAALRAP VTDMGNLPQN LFATRGAPPM LDADADDYLR RTV NAGNRL APVPVFGQML PQVPAGLARG QQSVCEFIAT PVSVDLAYFR RACNPRGRAA GEVHGEEGLM FDHSHADPAH PHRA TANPW ASQRHSYADR LYNGQYNMSG PAYSPCFKFF TPAEAVAKSR GLARLIADTG AAASPTSNGE YQFKRPVGAG ELVED PCAL FQEAYPPLCA SDSALLRTPL GAEEHFAQYL IRDESPLKGC FQHASA |
-Macromolecule #2: Triplex capsid protein 2
| Macromolecule | Name: Triplex capsid protein 2 / type: protein_or_peptide / ID: 2 / Number of copies: 10 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Suid alphaherpesvirus 1 Suid alphaherpesvirus 1 |
| Molecular weight | Theoretical: 31.763766 KDa |
| Sequence | String: MEVDIALPTL SPGDLSALQR CEGRVVFLET LRRHATLREV ALPCGGDVLA AMAAYRRRFA AVITRVTPHR MLATPLGVGG RGQSLVLQN TGPFDLTNGD HVCLVPPLLG DECLRLTSAN LELRFPMTLP LAQARELTAR VVARAAETLR GGAPARGADV V FSNGRRYQ ...String: MEVDIALPTL SPGDLSALQR CEGRVVFLET LRRHATLREV ALPCGGDVLA AMAAYRRRFA AVITRVTPHR MLATPLGVGG RGQSLVLQN TGPFDLTNGD HVCLVPPLLG DECLRLTSAN LELRFPMTLP LAQARELTAR VVARAAETLR GGAPARGADV V FSNGRRYQ LPPPHRDNAE AATRSLVLNM IFLLNEGAVI LLSLIPNLLT LGAQDGYANA VIQLGSATRE LGQLVRQPPP PL PQDHARR FCVFEALEAW IASASRLGDT LGTRPVARVC IFDGPPTVPP GEKAAVVEV |
-Macromolecule #3: Triplex capsid protein 1
| Macromolecule | Name: Triplex capsid protein 1 / type: protein_or_peptide / ID: 3 / Number of copies: 10 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Suid alphaherpesvirus 1 Suid alphaherpesvirus 1 |
| Molecular weight | Theoretical: 40.008594 KDa |
| Sequence | String: MSVQIGNGLL MVVAPGTLTV GSARARLIRQ VTLADFCEPQ AERPGLVVLA LRHPADLAGA AYAATPPGKN HRDLEEAWLA LDEGGRGLG GDGIRASVVS LNFLVAAAEN ADDALRAHVT TNYRDRRTAA RLERFATVLR AMIRSHVFPH RALHVLGGLL G HVTQDRLA ...String: MSVQIGNGLL MVVAPGTLTV GSARARLIRQ VTLADFCEPQ AERPGLVVLA LRHPADLAGA AYAATPPGKN HRDLEEAWLA LDEGGRGLG GDGIRASVVS LNFLVAAAEN ADDALRAHVT TNYRDRRTAA RLERFATVLR AMIRSHVFPH RALHVLGGLL G HVTQDRLA SVTCVARGDQ EAARTNDMAA RRSQVHVPAC ALMDVDRELR LGGDDGLRFA YLVFVYTQRH RREALRVHVA VS RLPELGD ALSFLLAGTR VDNAIHGTDE ADAPAAPAAA AAFPAYLFND PRSARCPTGR LNTPAAEALP VWAPDMRGRA TRN SCMYAA YVRLGTVERV VRRAERCGSV DLPLAHMERF TWDVGAWEEC FF |
-Macromolecule #4: Small capsomere-interacting protein
| Macromolecule | Name: Small capsomere-interacting protein / type: protein_or_peptide / ID: 4 / Number of copies: 15 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Suid alphaherpesvirus 1 Suid alphaherpesvirus 1 |
| Molecular weight | Theoretical: 11.509209 KDa |
| Sequence | String: MSFDPNNPRT ITAQTLEGAL PVDILLRLNR ATGLQMDAAE AHAIVEDARR TLFIGTSLAL VNLRRAHDKH LVERQPMFAT SDYSSWARP TVGLKRTFCP RPPP |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F30 |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Tecnai F30 / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 4.53 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 8899 |
|---|---|
| Initial angle assignment | Type: RANDOM ASSIGNMENT |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller