+ データを開く
データを開く
- 基本情報
基本情報
| 登録情報 |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| タイトル | Structure of mumps virus nucleoprotein without C-arm | |||||||||
 マップデータ マップデータ | MuV nucleocapsid with arm truncated | |||||||||
 試料 試料 |
| |||||||||
 キーワード キーワード | nucleocapsid / Mumps virus / NUCLEAR PROTEIN | |||||||||
| 機能・相同性 | Paramyxovirus nucleocapsid protein / Paramyxovirus nucleocapsid protein / helical viral capsid / viral nucleocapsid / host cell cytoplasm / ribonucleoprotein complex / structural molecule activity / RNA binding / Nucleoprotein 機能・相同性情報 機能・相同性情報 | |||||||||
| 生物種 |   Mumps virus (strain Jeryl-Lynn) (ウイルス) / Mumps virus (strain Jeryl-Lynn) (ウイルス) /  | |||||||||
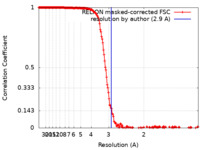
| 手法 | らせん対称体再構成法 / クライオ電子顕微鏡法 / 解像度: 2.9 Å | |||||||||
 データ登録者 データ登録者 | Shen Q / Shan H / Zhang N / Qin Y | |||||||||
| 資金援助 |  中国, 1件 中国, 1件
| |||||||||
 引用 引用 |  ジャーナル: Commun Biol / 年: 2021 ジャーナル: Commun Biol / 年: 2021タイトル: Structural plasticity of mumps virus nucleocapsids with cryo-EM structures. 著者: Hong Shan / Xin Su / Tianhao Li / Yuqi Qin / Na Zhang / Liuyan Yang / Linsha Ma / Yun Bai / Lei Qi / Yunhui Liu / Qing-Tao Shen /  要旨: Mumps virus (MuV) is a highly contagious human pathogen and frequently causes worldwide outbreaks despite available vaccines. Similar to other mononegaviruses such as Ebola and rabies, MuV uses a ...Mumps virus (MuV) is a highly contagious human pathogen and frequently causes worldwide outbreaks despite available vaccines. Similar to other mononegaviruses such as Ebola and rabies, MuV uses a single-stranded negative-sense RNA as its genome, which is enwrapped by viral nucleoproteins into the helical nucleocapsid. The nucleocapsid acts as a scaffold for genome condensation and as a template for RNA replication and transcription. Conformational changes in the MuV nucleocapsid are required to switch between different activities, but the underlying mechanism remains elusive due to the absence of high-resolution structures. Here, we report two MuV nucleoprotein-RNA rings with 13 and 14 protomers, one stacked-ring filament and two nucleocapsids with distinct helical pitches, in dense and hyperdense states, at near-atomic resolutions using cryo-electron microscopy. Structural analysis of these in vitro assemblies indicates that the C-terminal tail of MuV nucleoprotein likely regulates the assembly of helical nucleocapsids, and the C-terminal arm may be relevant for the transition between the dense and hyperdense states of helical nucleocapsids. Our results provide the molecular mechanism for structural plasticity among different MuV nucleocapsids and create a possible link between structural plasticity and genome condensation. | |||||||||
| 履歴 |
|
- 構造の表示
構造の表示
| 添付画像 |
|---|
- ダウンロードとリンク
ダウンロードとリンク
-EMDBアーカイブ
| マップデータ |  emd_31367.map.gz emd_31367.map.gz | 139.1 MB |  EMDBマップデータ形式 EMDBマップデータ形式 | |
|---|---|---|---|---|
| ヘッダ (付随情報) |  emd-31367-v30.xml emd-31367-v30.xml emd-31367.xml emd-31367.xml | 11.6 KB 11.6 KB | 表示 表示 |  EMDBヘッダ EMDBヘッダ |
| FSC (解像度算出) |  emd_31367_fsc.xml emd_31367_fsc.xml | 18.2 KB | 表示 |  FSCデータファイル FSCデータファイル |
| 画像 |  emd_31367.png emd_31367.png | 292.6 KB | ||
| Filedesc metadata |  emd-31367.cif.gz emd-31367.cif.gz | 5.6 KB | ||
| アーカイブディレクトリ |  http://ftp.pdbj.org/pub/emdb/structures/EMD-31367 http://ftp.pdbj.org/pub/emdb/structures/EMD-31367 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-31367 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-31367 | HTTPS FTP |
-検証レポート
| 文書・要旨 |  emd_31367_validation.pdf.gz emd_31367_validation.pdf.gz | 620.6 KB | 表示 |  EMDB検証レポート EMDB検証レポート |
|---|---|---|---|---|
| 文書・詳細版 |  emd_31367_full_validation.pdf.gz emd_31367_full_validation.pdf.gz | 620.2 KB | 表示 | |
| XML形式データ |  emd_31367_validation.xml.gz emd_31367_validation.xml.gz | 16 KB | 表示 | |
| CIF形式データ |  emd_31367_validation.cif.gz emd_31367_validation.cif.gz | 22 KB | 表示 | |
| アーカイブディレクトリ |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-31367 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-31367 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-31367 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-31367 | HTTPS FTP |
-関連構造データ
| 関連構造データ |  7exaMC  7ewqC M: このマップから作成された原子モデル C: 同じ文献を引用 ( |
|---|---|
| 類似構造データ | 類似検索 - 機能・相同性  F&H 検索 F&H 検索 |
- リンク
リンク
| EMDBのページ |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| 「今月の分子」の関連する項目 |
- マップ
マップ
| ファイル |  ダウンロード / ファイル: emd_31367.map.gz / 形式: CCP4 / 大きさ: 512 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) ダウンロード / ファイル: emd_31367.map.gz / 形式: CCP4 / 大きさ: 512 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | MuV nucleocapsid with arm truncated | ||||||||||||||||||||||||||||||||||||
| 投影像・断面図 | 画像のコントロール
画像は Spider により作成 | ||||||||||||||||||||||||||||||||||||
| ボクセルのサイズ | X=Y=Z: 0.66 Å | ||||||||||||||||||||||||||||||||||||
| 密度 |
| ||||||||||||||||||||||||||||||||||||
| 対称性 | 空間群: 1 | ||||||||||||||||||||||||||||||||||||
| 詳細 | EMDB XML:
|
-添付データ
- 試料の構成要素
試料の構成要素
-全体 : truncated mumps virus ncleocapsid
| 全体 | 名称: truncated mumps virus ncleocapsid |
|---|---|
| 要素 |
|
-超分子 #1: truncated mumps virus ncleocapsid
| 超分子 | 名称: truncated mumps virus ncleocapsid / タイプ: complex / ID: 1 / 親要素: 0 / 含まれる分子: all / 詳細: C-arm, C-tail removed |
|---|---|
| 由来(天然) | 生物種:  |
-分子 #1: Nucleoprotein
| 分子 | 名称: Nucleoprotein / タイプ: protein_or_peptide / ID: 1 / コピー数: 1 / 光学異性体: LEVO |
|---|---|
| 由来(天然) | 生物種:  Mumps virus (strain Jeryl-Lynn) (ウイルス) / 株: Jeryl-Lynn Mumps virus (strain Jeryl-Lynn) (ウイルス) / 株: Jeryl-Lynn |
| 分子量 | 理論値: 61.470008 KDa |
| 組換発現 | 生物種:  |
| 配列 | 文字列: MSSVLKAFER FTIEQELQDR GEEGSIPPET LKSAVKVFVI NTPNPTTRYQ MLNFCLRIIC SQNARASHRV GALITLFSLP SAGMQNHIR LADRSPEAQI ERCEIDGFEP GTYRLIPNAR ANLTANEIAA YALLADDLPP TINNGTPYVH ADVEGQPCDE I EQFLDRCY ...文字列: MSSVLKAFER FTIEQELQDR GEEGSIPPET LKSAVKVFVI NTPNPTTRYQ MLNFCLRIIC SQNARASHRV GALITLFSLP SAGMQNHIR LADRSPEAQI ERCEIDGFEP GTYRLIPNAR ANLTANEIAA YALLADDLPP TINNGTPYVH ADVEGQPCDE I EQFLDRCY SVLIQAWVMV CKCMTAYDQP AGSADRRFAK YQQQGRLEAR YMLQPEAQRL IQTAIRKSLV VRQYLTFELQ LA RRQGLLS NRYYAMVGDI GKYIENSGLT AFFLTLKYAL GTKWSPLSLA AFTGELTKLR SLMMLYRGLG EQARYLALLE APQ IMDFAP GGYPLIFSYA MGVGTVLDVQ MRNYTYARPF LNGYYFQIGV ETARRQQGTV DNRVADDLGL TPEQRTEVTQ LVDR LARGR GAGIPGGPVN PFVPPVQQQQ PAAVYEDIPA LEESDDDGDE DGGAGFQNGV QLPAVRQGGQ TDFRAQPLQD PIQAQ LFMP LYPQVSNMPN NQNHQINRIG GLEHQDLLRY NENGDSQQDA RGEHVNTFPN NPNQNAQLQV GDWDE UniProtKB: Nucleoprotein |
-分子 #2: RNA (5'-R(P*UP*UP*UP*UP*UP*U)-3')
| 分子 | 名称: RNA (5'-R(P*UP*UP*UP*UP*UP*U)-3') / タイプ: rna / ID: 2 / コピー数: 1 |
|---|---|
| 由来(天然) | 生物種:  |
| 分子量 | 理論値: 1.792037 KDa |
| 配列 | 文字列: UUUUUU |
-実験情報
-構造解析
| 手法 | クライオ電子顕微鏡法 |
|---|---|
 解析 解析 | らせん対称体再構成法 |
| 試料の集合状態 | helical array |
- 試料調製
試料調製
| 濃度 | 1 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 緩衝液 | pH: 7.4 構成要素:
| |||||||||
| グリッド | モデル: Quantifoil R2/1 / 材質: COPPER / 支持フィルム - 材質: CARBON / 支持フィルム - トポロジー: CONTINUOUS | |||||||||
| 凍結 | 凍結剤: ETHANE / チャンバー内湿度: 100 % / チャンバー内温度: 277 K / 装置: FEI VITROBOT MARK IV |
- 電子顕微鏡法
電子顕微鏡法
| 顕微鏡 | FEI TITAN KRIOS |
|---|---|
| 撮影 | フィルム・検出器のモデル: GATAN K3 (6k x 4k) / 平均電子線量: 40.0 e/Å2 |
| 電子線 | 加速電圧: 300 kV / 電子線源:  FIELD EMISSION GUN FIELD EMISSION GUN |
| 電子光学系 | 照射モード: FLOOD BEAM / 撮影モード: BRIGHT FIELD |
| 実験機器 |  モデル: Titan Krios / 画像提供: FEI Company |
 ムービー
ムービー コントローラー
コントローラー











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)