+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-30595 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Mycobacterium smegmatis Sdh1 complex in the apo form | ||||||||||||
 Map data Map data | |||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | succinate dehydrogenase / electron transport chain / Mycobacterium smegmatis / Sdh1 / SQR / OXIDOREDUCTASE | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationOxidoreductases; Acting on the CH-CH group of donors / membrane => GO:0016020 / tricarboxylic acid cycle / 2 iron, 2 sulfur cluster binding / 4 iron, 4 sulfur cluster binding / electron transfer activity / oxidoreductase activity / metal ion binding Similarity search - Function | ||||||||||||
| Biological species |  Mycolicibacterium smegmatis MC2 51 (bacteria) / Mycolicibacterium smegmatis MC2 51 (bacteria) /  Mycolicibacterium smegmatis (bacteria) Mycolicibacterium smegmatis (bacteria) | ||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.88 Å | ||||||||||||
 Authors Authors | Zhou X / Gao Y | ||||||||||||
| Funding support |  China, 3 items China, 3 items
| ||||||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2021 Journal: Proc Natl Acad Sci U S A / Year: 2021Title: Architecture of the mycobacterial succinate dehydrogenase with a membrane-embedded Rieske FeS cluster. Authors: Xiaoting Zhou / Yan Gao / Weiwei Wang / Xiaolin Yang / Xiuna Yang / Fengjiang Liu / Yanting Tang / Sin Man Lam / Guanghou Shui / Lu Yu / Changlin Tian / Luke W Guddat / Quan Wang / Zihe Rao / Hongri Gong /   Abstract: Complex II, also known as succinate dehydrogenase (SQR) or fumarate reductase (QFR), is an enzyme involved in both the Krebs cycle and oxidative phosphorylation. Mycobacterial Sdh1 has recently been ...Complex II, also known as succinate dehydrogenase (SQR) or fumarate reductase (QFR), is an enzyme involved in both the Krebs cycle and oxidative phosphorylation. Mycobacterial Sdh1 has recently been identified as a new class of respiratory complex II (type F) but with an unknown electron transfer mechanism. Here, using cryoelectron microscopy, we have determined the structure of Sdh1 in the presence and absence of the substrate, ubiquinone-1, at 2.53-Å and 2.88-Å resolution, respectively. Sdh1 comprises three subunits, two that are water soluble, SdhA and SdhB, and one that is membrane spanning, SdhC. Within these subunits we identified a quinone-binding site and a rarely observed Rieske-type [2Fe-2S] cluster, the latter being embedded in the transmembrane region. A mutant, where two His ligands of the Rieske-type [2Fe-2S] were changed to alanine, abolished the quinone reduction activity of the Sdh1. Our structures allow the proposal of an electron transfer pathway that connects the substrate-binding and quinone-binding sites. Given the unique features of Sdh1 and its essential role in , these structures will facilitate antituberculosis drug discovery efforts that specifically target this complex. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_30595.map.gz emd_30595.map.gz | 203.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-30595-v30.xml emd-30595-v30.xml emd-30595.xml emd-30595.xml | 14.8 KB 14.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_30595.png emd_30595.png | 127.9 KB | ||
| Filedesc metadata |  emd-30595.cif.gz emd-30595.cif.gz | 6.2 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-30595 http://ftp.pdbj.org/pub/emdb/structures/EMD-30595 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-30595 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-30595 | HTTPS FTP |
-Validation report
| Summary document |  emd_30595_validation.pdf.gz emd_30595_validation.pdf.gz | 503.1 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_30595_full_validation.pdf.gz emd_30595_full_validation.pdf.gz | 502.7 KB | Display | |
| Data in XML |  emd_30595_validation.xml.gz emd_30595_validation.xml.gz | 7.1 KB | Display | |
| Data in CIF |  emd_30595_validation.cif.gz emd_30595_validation.cif.gz | 8.1 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-30595 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-30595 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-30595 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-30595 | HTTPS FTP |
-Related structure data
| Related structure data |  7d6xMC  7d6vC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_30595.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_30595.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.82 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
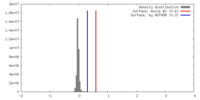
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Mycobacterium smegmatis Sdh1 complex in apo form
| Entire | Name: Mycobacterium smegmatis Sdh1 complex in apo form |
|---|---|
| Components |
|
-Supramolecule #1: Mycobacterium smegmatis Sdh1 complex in apo form
| Supramolecule | Name: Mycobacterium smegmatis Sdh1 complex in apo form / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 / Details: succinate dehydrogenase |
|---|---|
| Source (natural) | Organism:  Mycolicibacterium smegmatis MC2 51 (bacteria) Mycolicibacterium smegmatis MC2 51 (bacteria) |
-Macromolecule #1: Succinate dehydrogenase subunit A
| Macromolecule | Name: Succinate dehydrogenase subunit A / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO EC number: Oxidoreductases; Acting on the CH-CH group of donors |
|---|---|
| Source (natural) | Organism:  Mycolicibacterium smegmatis (bacteria) Mycolicibacterium smegmatis (bacteria) |
| Molecular weight | Theoretical: 70.14718 KDa |
| Sequence | String: MSELERHSYD VVVIGAGGAG LRAVIEARER GLRVAVVTKS LFGKAHTVMA EGGCAAAMRN VNTKDSWQVH FGDTMRGGKF LNNWRMAEL HAQEAPDRVW ELETYGALFD RTKDGKISQR NFGGHTYPRL AHVGDRTGLE IIRTLQQKIV SLQQEDKREL G DYEARIRV ...String: MSELERHSYD VVVIGAGGAG LRAVIEARER GLRVAVVTKS LFGKAHTVMA EGGCAAAMRN VNTKDSWQVH FGDTMRGGKF LNNWRMAEL HAQEAPDRVW ELETYGALFD RTKDGKISQR NFGGHTYPRL AHVGDRTGLE IIRTLQQKIV SLQQEDKREL G DYEARIRV FHETSITELI LDDGKIAGAF GYYRETGNFV LFEAPAVVLA TGGIGKSFKV SSNSWEYTGD GHALALRAGS AL INMEFIQ FHPTGMVWPL SVKGILVTEG VRGDGGVLKN SEGKRFMFDY IPSVFKGQYA ETEEEADQWL KDNDSARRTP DLL PRDEVA RAINAEVKAG RGSPHGGVYL DIASRMPAEE IKRRLPSMYH QFIELAEVDI TKDAMEVGPT CHYVMGGIEV DPDT AAGAT PGLFAAGECS GGMHGSNRLG GNSLSDLLVF GRRAGLGAAD YVRALPDRPK VSEAAVEDAT RLVLAPFEPK AEPEN PYTL HAELQQSMND LVGIIRKEAE IQEALDRLQE LKRRYANVTV EGGRVFNPGW HLAIDMRNML LVSECVAKAA LQRTES RGG HTRDDYPEMD ANWRNTLLVC RVSGGDPVVP DVTVTPEQQV PMRPDLLGCF ELSELEKYYT PEELAEHPER KG UniProtKB: Succinate dehydrogenase subunit A |
-Macromolecule #2: Fumarate reductase iron-sulfur subunit
| Macromolecule | Name: Fumarate reductase iron-sulfur subunit / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Mycolicibacterium smegmatis (bacteria) Mycolicibacterium smegmatis (bacteria) |
| Molecular weight | Theoretical: 28.851988 KDa |
| Sequence | String: MATYDAKLRV WRGDDTGGEL HDYTVEVNDG EVVLDIIHRL QATQTPDLAV RWNCKAGKCG SCSAEINGRP RLMCMTRMST FGEDEVVTV TPLRTFPVMR DLVTDVSFNY EKARQIPSFT PPKDLQPGEY RMQQEDVNRS QEFRKCIECF LCQNVCHVVR D HEENKENF ...String: MATYDAKLRV WRGDDTGGEL HDYTVEVNDG EVVLDIIHRL QATQTPDLAV RWNCKAGKCG SCSAEINGRP RLMCMTRMST FGEDEVVTV TPLRTFPVMR DLVTDVSFNY EKARQIPSFT PPKDLQPGEY RMQQEDVNRS QEFRKCIECF LCQNVCHVVR D HEENKENF AGPRFHMRIA ELDMHPLDTV DRKEMAQDEF GLGYCNITKC CTEVCPEHIK ITDNALIPMK ERVADRKYDP IV WLGNKLF RR UniProtKB: Fumarate reductase iron-sulfur subunit |
-Macromolecule #3: Succinate dehydrogenase (Membrane anchor subunit)
| Macromolecule | Name: Succinate dehydrogenase (Membrane anchor subunit) / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Mycolicibacterium smegmatis (bacteria) Mycolicibacterium smegmatis (bacteria) |
| Molecular weight | Theoretical: 32.583545 KDa |
| Sequence | String: MSAPTADRRA TGVFSPRRAQ IPERTLRTDR WWQAPLLTNL GLAAFVIYAT IRAFWGSAYW VADYHYLTPF YSPCVSTACA PGSSHFGQW VGDLPWFIPM AFISLPFLLA FRLTCYYYRK AYYRSVWQSP TACAVAEPHA KYTGETRFPL ILQNIHRYFF Y AAVLISLV ...String: MSAPTADRRA TGVFSPRRAQ IPERTLRTDR WWQAPLLTNL GLAAFVIYAT IRAFWGSAYW VADYHYLTPF YSPCVSTACA PGSSHFGQW VGDLPWFIPM AFISLPFLLA FRLTCYYYRK AYYRSVWQSP TACAVAEPHA KYTGETRFPL ILQNIHRYFF Y AAVLISLV NTYDAITAFH SPSGFGFGLG NVILTGNVIL LWVYTLSCHS CRHVTGGRLK HFSKHPVRYW IWTQVSKLNT RH MLFAWIT LGTLVLTDFY IMLVASGTIS DLRFIGHHHH HHHHHH UniProtKB: Succinate dehydrogenase (Membrane anchor subunit) |
-Macromolecule #4: FLAVIN-ADENINE DINUCLEOTIDE
| Macromolecule | Name: FLAVIN-ADENINE DINUCLEOTIDE / type: ligand / ID: 4 / Number of copies: 1 / Formula: FAD |
|---|---|
| Molecular weight | Theoretical: 785.55 Da |
| Chemical component information |  ChemComp-FAD: |
-Macromolecule #5: FE2/S2 (INORGANIC) CLUSTER
| Macromolecule | Name: FE2/S2 (INORGANIC) CLUSTER / type: ligand / ID: 5 / Number of copies: 2 / Formula: FES |
|---|---|
| Molecular weight | Theoretical: 175.82 Da |
| Chemical component information |  ChemComp-FES: |
-Macromolecule #6: IRON/SULFUR CLUSTER
| Macromolecule | Name: IRON/SULFUR CLUSTER / type: ligand / ID: 6 / Number of copies: 1 / Formula: SF4 |
|---|---|
| Molecular weight | Theoretical: 351.64 Da |
| Chemical component information |  ChemComp-FS1: |
-Macromolecule #7: FE3-S4 CLUSTER
| Macromolecule | Name: FE3-S4 CLUSTER / type: ligand / ID: 7 / Number of copies: 1 / Formula: F3S |
|---|---|
| Molecular weight | Theoretical: 295.795 Da |
| Chemical component information |  ChemComp-F3S: |
-Macromolecule #8: (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY...
| Macromolecule | Name: (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE type: ligand / ID: 8 / Number of copies: 1 / Formula: PEV |
|---|---|
| Molecular weight | Theoretical: 720.012 Da |
| Chemical component information |  ChemComp-PEV: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 5 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 281 K / Instrument: FEI VITROBOT MARK II |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Detector mode: SUPER-RESOLUTION / Digitization - Frames/image: 2-40 / Average exposure time: 5.0 sec. / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 2.88 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 254341 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller

























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)