[English] 日本語
 Yorodumi
Yorodumi- EMDB-30234: Mycobacterium smegmatis arabinosyltransferase complex EmbB2-AcpM2... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-30234 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Mycobacterium smegmatis arabinosyltransferase complex EmbB2-AcpM2 in substrate DPA bound asymmetric "active state | |||||||||
 Map data Map data | half map B from cryoSPARC | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Mycobacterium tuberculosis / EmbB / cryo-EM / ethambutol / cell wall synthesis / arabinoglacatan / arabinosyltransferase / acyl-carrier-protein / TRANSFERASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationindolylacetylinositol arabinosyltransferase / indolylacetylinositol arabinosyltransferase activity / arabinosyltransferase activity / Actinobacterium-type cell wall biogenesis / Transferases; Glycosyltransferases; Pentosyltransferases / acyl carrier activity / cell wall organization / plasma membrane / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Mycolicibacterium smegmatis MC2 155 (bacteria) Mycolicibacterium smegmatis MC2 155 (bacteria) | |||||||||
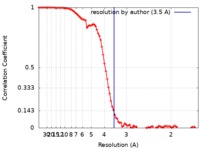
| Method | single particle reconstruction / cryo EM / Resolution: 3.5 Å | |||||||||
 Authors Authors | Gao RG / Zhang L | |||||||||
| Funding support |  China, 2 items China, 2 items
| |||||||||
 Citation Citation |  Journal: Protein Cell / Year: 2020 Journal: Protein Cell / Year: 2020Title: Cryo-EM snapshots of mycobacterial arabinosyltransferase complex EmbB-AcpM. Authors: Lu Zhang / Yao Zhao / Ruogu Gao / Jun Li / Xiuna Yang / Yan Gao / Wei Zhao / Sudagar S Gurcha / Natacha Veerapen / Sarah M Batt / Kajelle Kaur Besra / Wenqing Xu / Lijun Bi / Xian'en Zhang / ...Authors: Lu Zhang / Yao Zhao / Ruogu Gao / Jun Li / Xiuna Yang / Yan Gao / Wei Zhao / Sudagar S Gurcha / Natacha Veerapen / Sarah M Batt / Kajelle Kaur Besra / Wenqing Xu / Lijun Bi / Xian'en Zhang / Luke W Guddat / Haitao Yang / Quan Wang / Gurdyal S Besra / Zihe Rao /    Abstract: Inhibition of Mycobacterium tuberculosis (Mtb) cell wall assembly is an established strategy for anti-TB chemotherapy. Arabinosyltransferase EmbB, which catalyzes the transfer of arabinose from the ...Inhibition of Mycobacterium tuberculosis (Mtb) cell wall assembly is an established strategy for anti-TB chemotherapy. Arabinosyltransferase EmbB, which catalyzes the transfer of arabinose from the donor decaprenyl-phosphate-arabinose (DPA) to its arabinosyl acceptor is an essential enzyme for Mtb cell wall synthesis. Analysis of drug resistance mutations suggests that EmbB is the main target of the front-line anti-TB drug, ethambutol. Herein, we report the cryo-EM structures of Mycobacterium smegmatis EmbB in its "resting state" and DPA-bound "active state". EmbB is a fifteen-transmembrane-spanning protein, assembled as a dimer. Each protomer has an associated acyl-carrier-protein (AcpM) on their cytoplasmic surface. Conformational changes upon DPA binding indicate an asymmetric movement within the EmbB dimer during catalysis. Functional studies have identified critical residues in substrate recognition and catalysis, and demonstrated that ethambutol inhibits transferase activity of EmbB by competing with DPA. The structures represent the first step directed towards a rational approach for anti-TB drug discovery. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_30234.map.gz emd_30234.map.gz | 59.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-30234-v30.xml emd-30234-v30.xml emd-30234.xml emd-30234.xml | 20.5 KB 20.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_30234_fsc.xml emd_30234_fsc.xml | 8.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_30234.png emd_30234.png | 136.5 KB | ||
| Masks |  emd_30234_msk_1.map emd_30234_msk_1.map | 64 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-30234.cif.gz emd-30234.cif.gz | 6.5 KB | ||
| Others |  emd_30234_additional.map.gz emd_30234_additional.map.gz emd_30234_half_map_1.map.gz emd_30234_half_map_1.map.gz emd_30234_half_map_2.map.gz emd_30234_half_map_2.map.gz | 28.8 MB 59.4 MB 59.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-30234 http://ftp.pdbj.org/pub/emdb/structures/EMD-30234 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-30234 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-30234 | HTTPS FTP |
-Validation report
| Summary document |  emd_30234_validation.pdf.gz emd_30234_validation.pdf.gz | 1.2 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_30234_full_validation.pdf.gz emd_30234_full_validation.pdf.gz | 1.2 MB | Display | |
| Data in XML |  emd_30234_validation.xml.gz emd_30234_validation.xml.gz | 16.3 KB | Display | |
| Data in CIF |  emd_30234_validation.cif.gz emd_30234_validation.cif.gz | 20.8 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-30234 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-30234 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-30234 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-30234 | HTTPS FTP |
-Related structure data
| Related structure data |  7bwrMC  7bx8C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_30234.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_30234.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map B from cryoSPARC | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.82 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
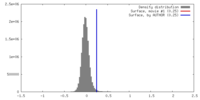
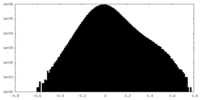
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_30234_msk_1.map emd_30234_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
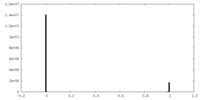
| Density Histograms |
-Additional map: mask used in refinement from cryoSPARC
| File | emd_30234_additional.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | mask used in refinement from cryoSPARC | ||||||||||||
| Projections & Slices |
| ||||||||||||
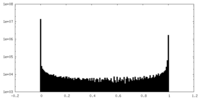
| Density Histograms |
-Half map: half map A from cryoSPARC
| File | emd_30234_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map A from cryoSPARC | ||||||||||||
| Projections & Slices |
| ||||||||||||
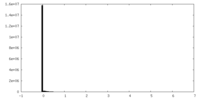
| Density Histograms |
-Half map: half map B from cryoSPARC
| File | emd_30234_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map B from cryoSPARC | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Mycobacterial Arabinosyltransferase Complex EmbB2-AcpM2
| Entire | Name: Mycobacterial Arabinosyltransferase Complex EmbB2-AcpM2 |
|---|---|
| Components |
|
-Supramolecule #1: Mycobacterial Arabinosyltransferase Complex EmbB2-AcpM2
| Supramolecule | Name: Mycobacterial Arabinosyltransferase Complex EmbB2-AcpM2 type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  Mycolicibacterium smegmatis MC2 155 (bacteria) Mycolicibacterium smegmatis MC2 155 (bacteria) |
| Molecular weight | Theoretical: 254 kDa/nm |
-Macromolecule #1: Integral membrane indolylacetylinositol arabinosyltransferase EmbB
| Macromolecule | Name: Integral membrane indolylacetylinositol arabinosyltransferase EmbB type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO / EC number: indolylacetylinositol arabinosyltransferase |
|---|---|
| Source (natural) | Organism:  Mycolicibacterium smegmatis MC2 155 (bacteria) Mycolicibacterium smegmatis MC2 155 (bacteria) |
| Molecular weight | Theoretical: 116.870289 KDa |
| Recombinant expression | Organism:  Mycolicibacterium smegmatis MC2 155 (bacteria) Mycolicibacterium smegmatis MC2 155 (bacteria) |
| Sequence | String: MSGNMDEAVS GNMDEAVSAG KDVRIARWVA TIAGLLGFVL SVSIPLLPVT QTTATLNWPQ QGRLDNVTAP LISQAPLELT ATVPCSVVR DLPPEGGLVF GTAPAEGRDA ALNAMLVNVT ETRVDVIVRN VVVASVNRDR VAGPDCQRIE ITSNLDGTYA D FVGLTQIS ...String: MSGNMDEAVS GNMDEAVSAG KDVRIARWVA TIAGLLGFVL SVSIPLLPVT QTTATLNWPQ QGRLDNVTAP LISQAPLELT ATVPCSVVR DLPPEGGLVF GTAPAEGRDA ALNAMLVNVT ETRVDVIVRN VVVASVNRDR VAGPDCQRIE ITSNLDGTYA D FVGLTQIS GEDAGKLQRT GYPDPNLRPA IVGVFTDLTG PAPQGLSVSA EIDTRFTTHP TALKLAAMLL AIVSTVIALL AL WRLDRLD GRRMHRLIPT RWRTVTAVDG VVVGGMAIWY VIGANSSDDG YILQMARTAE HAGYMANYFR WFGSPEDPFG WYY NVLALM TKVSDASIWI RLPDLICALI CWLLLSREVL PRLGPAVAGS RAAMWAAGLV LLGAWMPFNN GLRPEGQIAT GALI TYVLI ERAVTSGRLT PAALAITTAA FTLGIQPTGL IAVAALLAGG RPILRIVMRR RRLVGTWPLI APLLAAGTVI LAVVF ADQT IATVLEATRI RTAIGPSQEW WTENLRYYYL ILPTTDGAIS RRVAFVFTAM CLFPSLFMML RRKHIAGVAR GPAWRL MGI IFATMFFLMF TPTKWIHHFG LFAAVGGAMA ALATVLVSPT VLRSARNRMA FLSLVLFVLA FCFASTNGWW YVSNFGA PF NNSVPKVGGV QISAIFFALS AIAALWAFWL HLTRRTESRV VDRLTAAPIP VAAGFMVVVM MASMAIGVVR QYPTYSNG W ANIRAFAGGC GLADDVLVEP DSNAGFLTPL PGAYGPLGPL GGEDPQGFSP DGVPDRIIAE AIRLNNPQPG TDYDWNRPI KLDEPGINGS TVPLPYGLDP KRVPVAGTYS TEAQQESRLS SAWYELPARD ETERAAHPLV VITAAGTITG ESVANGLTTG QTVDLEYAT RGPDGTLVPA GRVTPYDVGP TPSWRNLRYP RSEIPDDAVA VRVVAEDLSL SQGDWIAVTP PRVPELQSVQ E YVGSDQPV LMDWAVGLAF PCQQPMLHAN GVTEVPKFRI SPDYYAKLQS TDTWQDGING GLLGITDLLL RASVMSTYLS QD WGQDWGS LRKFDTVVEA TPAELDFGSQ THSGLYSPGP LRIRP UniProtKB: Integral membrane indolylacetylinositol arabinosyltransferase EmbB |
-Macromolecule #2: Meromycolate extension acyl carrier protein
| Macromolecule | Name: Meromycolate extension acyl carrier protein / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Mycolicibacterium smegmatis MC2 155 (bacteria) Mycolicibacterium smegmatis MC2 155 (bacteria) |
| Molecular weight | Theoretical: 10.743876 KDa |
| Recombinant expression | Organism:  Mycolicibacterium smegmatis MC2 155 (bacteria) Mycolicibacterium smegmatis MC2 155 (bacteria) |
| Sequence | String: MAATQEEIIA GLAEIIEEVT GIEPSEVTPE KSFVDDLDID SLSMVEIAVQ TEDKYGVKIP DEDLAGLRTV GDVVAYIQKL EEENPEAAA ALREKFAADQ UniProtKB: Meromycolate extension acyl carrier protein |
-Macromolecule #3: CALCIUM ION
| Macromolecule | Name: CALCIUM ION / type: ligand / ID: 3 / Number of copies: 1 / Formula: CA |
|---|---|
| Molecular weight | Theoretical: 40.078 Da |
-Macromolecule #4: [(2Z,6E,10E,14Z,18E,22Z,26Z)-3,7,11,15,19,23,27,31,35,39-decameth...
| Macromolecule | Name: [(2Z,6E,10E,14Z,18E,22Z,26Z)-3,7,11,15,19,23,27,31,35,39-decamethyltetraconta-2,6,10,14,18,22,26,30,34,38-decaenyl] [(2S,3S,4S,5R)-5-(hydroxymethyl)-3,4-bis(oxidanyl)oxolan-2-yl] hydrogen phosphate type: ligand / ID: 4 / Number of copies: 1 / Formula: F8L |
|---|---|
| Molecular weight | Theoretical: 911.28 Da |
| Chemical component information | 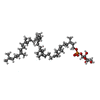 ChemComp-F8L: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)