[English] 日本語
 Yorodumi
Yorodumi- EMDB-29983: Cryo-EM structure of human TRPV1 in complex with the analgesic dr... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
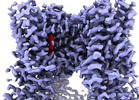
| Title | Cryo-EM structure of human TRPV1 in complex with the analgesic drug SB-366791 | ||||||||||||||||||
 Map data Map data | |||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationchemosensory behavior / temperature-gated ion channel activity / response to capsazepine / negative regulation of establishment of blood-brain barrier / sensory perception of mechanical stimulus / peptide secretion / detection of chemical stimulus involved in sensory perception of pain / cellular response to temperature stimulus / smooth muscle contraction involved in micturition / cellular response to acidic pH ...chemosensory behavior / temperature-gated ion channel activity / response to capsazepine / negative regulation of establishment of blood-brain barrier / sensory perception of mechanical stimulus / peptide secretion / detection of chemical stimulus involved in sensory perception of pain / cellular response to temperature stimulus / smooth muscle contraction involved in micturition / cellular response to acidic pH / excitatory extracellular ligand-gated monoatomic ion channel activity / thermoception / fever generation / detection of temperature stimulus involved in thermoception / glutamate secretion / negative regulation of systemic arterial blood pressure / chloride channel regulator activity / dendritic spine membrane / TRP channels / extracellular ligand-gated monoatomic ion channel activity / cellular response to ATP / negative regulation of heart rate / cellular response to alkaloid / behavioral response to pain / diet induced thermogenesis / intracellularly gated calcium channel activity / detection of temperature stimulus involved in sensory perception of pain / negative regulation of mitochondrial membrane potential / calcium ion import across plasma membrane / voltage-gated calcium channel activity / phosphatidylinositol binding / cellular response to nerve growth factor stimulus / calcium ion transmembrane transport / phosphoprotein binding / microglial cell activation / calcium channel activity / lipid metabolic process / response to peptide hormone / transmembrane signaling receptor activity / positive regulation of nitric oxide biosynthetic process / cellular response to tumor necrosis factor / cellular response to heat / positive regulation of cytosolic calcium ion concentration / postsynaptic membrane / protein homotetramerization / cell surface receptor signaling pathway / calmodulin binding / positive regulation of apoptotic process / external side of plasma membrane / neuronal cell body / negative regulation of transcription by RNA polymerase II / ATP binding / identical protein binding / membrane / metal ion binding / plasma membrane Similarity search - Function | ||||||||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.29 Å | ||||||||||||||||||
 Authors Authors | Neuberger A / Nadezhdin KD / Sobolevsky AI | ||||||||||||||||||
| Funding support |  United States, United States,  Germany, 5 items Germany, 5 items
| ||||||||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Human TRPV1 structure and inhibition by the analgesic SB-366791. Authors: Arthur Neuberger / Mai Oda / Yury A Nikolaev / Kirill D Nadezhdin / Elena O Gracheva / Sviatoslav N Bagriantsev / Alexander I Sobolevsky /  Abstract: Pain therapy has remained conceptually stagnant since the opioid crisis, which highlighted the dangers of treating pain with opioids. An alternative addiction-free strategy to conventional painkiller- ...Pain therapy has remained conceptually stagnant since the opioid crisis, which highlighted the dangers of treating pain with opioids. An alternative addiction-free strategy to conventional painkiller-based treatment is targeting receptors at the origin of the pain pathway, such as transient receptor potential (TRP) ion channels. Thus, a founding member of the vanilloid subfamily of TRP channels, TRPV1, represents one of the most sought-after pain therapy targets. The need for selective TRPV1 inhibitors extends beyond pain treatment, to other diseases associated with this channel, including psychiatric disorders. Here we report the cryo-electron microscopy structures of human TRPV1 in the apo state and in complex with the TRPV1-specific nanomolar-affinity analgesic antagonist SB-366791. SB-366791 binds to the vanilloid site and acts as an allosteric hTRPV1 inhibitor. SB-366791 binding site is supported by mutagenesis combined with electrophysiological recordings and can be further explored to design new drugs targeting TRPV1 in disease conditions. | ||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_29983.map.gz emd_29983.map.gz | 117.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-29983-v30.xml emd-29983-v30.xml emd-29983.xml emd-29983.xml | 21.5 KB 21.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_29983.png emd_29983.png | 174.7 KB | ||
| Others |  emd_29983_half_map_1.map.gz emd_29983_half_map_1.map.gz emd_29983_half_map_2.map.gz emd_29983_half_map_2.map.gz | 115.7 MB 115.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-29983 http://ftp.pdbj.org/pub/emdb/structures/EMD-29983 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29983 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29983 | HTTPS FTP |
-Validation report
| Summary document |  emd_29983_validation.pdf.gz emd_29983_validation.pdf.gz | 962.5 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_29983_full_validation.pdf.gz emd_29983_full_validation.pdf.gz | 962.1 KB | Display | |
| Data in XML |  emd_29983_validation.xml.gz emd_29983_validation.xml.gz | 13.9 KB | Display | |
| Data in CIF |  emd_29983_validation.cif.gz emd_29983_validation.cif.gz | 16.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29983 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29983 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29983 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29983 | HTTPS FTP |
-Related structure data
| Related structure data |  8gfaMC  8gf8C  8gf9C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_29983.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_29983.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 0.645 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_29983_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
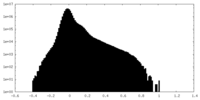
| Projections & Slices |
| ||||||||||||
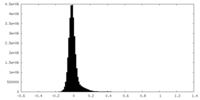
| Density Histograms |
-Half map: #1
| File | emd_29983_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
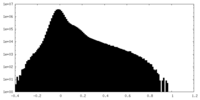
| Projections & Slices |
| ||||||||||||
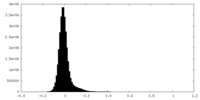
| Density Histograms |
- Sample components
Sample components
-Entire : sample 1
| Entire | Name: sample 1 |
|---|---|
| Components |
|
-Supramolecule #1: sample 1
| Supramolecule | Name: sample 1 / type: complex / ID: 1 / Chimera: Yes / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 94.97 KDa |
-Macromolecule #1: Transient receptor potential cation channel subfamily V member 1
| Macromolecule | Name: Transient receptor potential cation channel subfamily V member 1 type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 124.575281 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MTSKKWSSTD LGAAADPLQK DTCPDPLDGD PNSRPPPAKP QLSTAKSRTR LFGKGDSEEA FPVDCPHEEG ELDSCPTITV SPVITIQRP GDGPTGARLL SQDSVAASTE KTLRLYDRRS IFEAVAQNNC QDLESLLLFL QKSKKHLTDN EFKDPETGKT C LLKAMLNL ...String: MTSKKWSSTD LGAAADPLQK DTCPDPLDGD PNSRPPPAKP QLSTAKSRTR LFGKGDSEEA FPVDCPHEEG ELDSCPTITV SPVITIQRP GDGPTGARLL SQDSVAASTE KTLRLYDRRS IFEAVAQNNC QDLESLLLFL QKSKKHLTDN EFKDPETGKT C LLKAMLNL HDGQNTTIPL LLEIARQTDS LKELVNASYT DSYYKGQTAL HIAIERRNMA LVTLLVENGA DVQAAAHGDF FK KTKGRPG FYFGELPLSL AACTNQLGIV KFLLQNSWQT ADISARDSVG NTVLHALVEV ADNTADNTKF VTSMYNEILM LGA KLHPTL KLEELTNKKG MTPLALAAGT GKIGVLAYIL QREIQEPECR HLSRKFTEWA YGPVHSSLYD LSCIDTCEKN SVLE VIAYS SSETPNRHDM LLVEPLNRLL QDKWDRFVKR IFYFNFLVYC LYMIIFTMAA YYRPVDGLPP FKMEKTGDYF RVTGE ILSV LGGVYFFFRG IQYFLQRRPS MKTLFVDSYS EMLFFLQSLF MLATVVLYFS HLKEYVASMV FSLALGWTNM LYYTRG FQQ MGIYAVMIEK MILRDLCRFM FVYIVFLFGF STAVVTLIED GKNDSLPSES TSHRWRGPAC RPPDSSYNSL YSTCLEL FK FTIGMGDLEF TENYDFKAVF IILLLAYVIL TYILLLNMLI ALMGETVNKI AQESKNIWKL QRAITILDTE KSFLKCMR K AFRSGKLLQV GYTPDGKDDY RWCFRVDEVN WTTWNTNVGI INEDPGNCEG VKRTLSFSLR SSRVSGRHWK NFALVPLLR EASARDRQSA QPEEVYLRQF SGSLKPEDAE VFKSPAASGE KLVPRGSAAA AVSKGEELFT GVVPILVELD GDVNGHKFSV SGEGEGDAT YGKLTLKFIC TTGKLPVPWP TLVTTLTYGV QCFSRYPDHM KQHDFFKSAM PEGYVQERTI FFKDDGNYKT R AEVKFEGD TLVNRIELKG IDFKEDGNIL GHKLEYNYNS HNVYIMADKQ KNGIKVNFKI RHNIEDGSVQ LADHYQQNTP IG DGPVLLP DNHYLSTQSK LSKDPNEKRD HMVLLEFVTA AGITLGMDEL YKSGLRSWSH PQFEK |
-Macromolecule #2: (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(tri...
| Macromolecule | Name: (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate type: ligand / ID: 2 / Number of copies: 36 / Formula: POV |
|---|---|
| Molecular weight | Theoretical: 760.076 Da |
| Chemical component information |  ChemComp-POV: |
-Macromolecule #3: (2E)-3-(4-chlorophenyl)-N-(3-methoxyphenyl)prop-2-enamide
| Macromolecule | Name: (2E)-3-(4-chlorophenyl)-N-(3-methoxyphenyl)prop-2-enamide type: ligand / ID: 3 / Number of copies: 4 / Formula: ZEI |
|---|---|
| Molecular weight | Theoretical: 287.741 Da |
| Chemical component information | 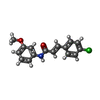 ChemComp-ZEI: |
-Macromolecule #4: 2-[2-[(1~{S},2~{S},4~{S},5'~{R},6~{R},7~{S},8~{R},9~{S},12~{S},13...
| Macromolecule | Name: 2-[2-[(1~{S},2~{S},4~{S},5'~{R},6~{R},7~{S},8~{R},9~{S},12~{S},13~{R},16~{S})-5',7,9,13-tetramethylspiro[5-oxapentacyclo[10.8.0.0^{2,9}.0^{4,8}.0^{13,18}]icos-18-ene-6,2'-oxane]-16-yl]oxyethyl]propane-1,3-diol type: ligand / ID: 4 / Number of copies: 4 / Formula: DU0 |
|---|---|
| Molecular weight | Theoretical: 516.752 Da |
| Chemical component information |  ChemComp-DU0: |
-Macromolecule #5: SODIUM ION
| Macromolecule | Name: SODIUM ION / type: ligand / ID: 5 / Number of copies: 2 |
|---|---|
| Molecular weight | Theoretical: 22.99 Da |
-Macromolecule #6: water
| Macromolecule | Name: water / type: ligand / ID: 6 / Number of copies: 105 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 3 mg/mL | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| ||||||||||||||||||
| Grid | Model: UltrAuFoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Support film - Material: GOLD / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE | ||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV | ||||||||||||||||||
| Details | human TRPV1 |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Digitization - Dimensions - Width: 5760 pixel / Digitization - Dimensions - Height: 4092 pixel / Number grids imaged: 1 / Number real images: 4188 / Average exposure time: 1.87 sec. / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 1.5 µm / Nominal defocus min: 0.75 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Software | Name:  Coot (ver. 0.9.8.1) Coot (ver. 0.9.8.1) |
|---|---|
| Refinement | Space: REAL |
| Output model |  PDB-8gfa: |
 Movie
Movie Controller
Controller






 Z
Z Y
Y X
X