+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | X-31 hemagglutinin in complex with FL-1061 Fab | ||||||||||||
 Map data Map data | half map 1 | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | influenza / fusogen / antibody / VIRAL PROTEIN | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationviral budding from plasma membrane / clathrin-dependent endocytosis of virus by host cell / host cell surface receptor binding / apical plasma membrane / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / virion attachment to host cell / host cell plasma membrane / virion membrane Similarity search - Function | ||||||||||||
| Biological species |   Influenza A virus / Influenza A virus /  | ||||||||||||
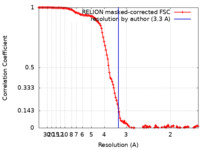
| Method | single particle reconstruction / cryo EM / Resolution: 3.3 Å | ||||||||||||
 Authors Authors | Windsor IW / Thornlow D / Schmidt AG | ||||||||||||
| Funding support |  United States, 3 items United States, 3 items
| ||||||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Antibodies elicited by a hyperglycoslated immunogen Authors: Windsor IW / Schmidt AG | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_29737.map.gz emd_29737.map.gz | 166.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-29737-v30.xml emd-29737-v30.xml emd-29737.xml emd-29737.xml | 16.8 KB 16.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_29737_fsc.xml emd_29737_fsc.xml | 12.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_29737.png emd_29737.png | 116.9 KB | ||
| Filedesc metadata |  emd-29737.cif.gz emd-29737.cif.gz | 6.2 KB | ||
| Others |  emd_29737_half_map_1.map.gz emd_29737_half_map_1.map.gz emd_29737_half_map_2.map.gz emd_29737_half_map_2.map.gz | 140.8 MB 140.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-29737 http://ftp.pdbj.org/pub/emdb/structures/EMD-29737 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29737 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29737 | HTTPS FTP |
-Validation report
| Summary document |  emd_29737_validation.pdf.gz emd_29737_validation.pdf.gz | 1007.6 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_29737_full_validation.pdf.gz emd_29737_full_validation.pdf.gz | 1007.2 KB | Display | |
| Data in XML |  emd_29737_validation.xml.gz emd_29737_validation.xml.gz | 20 KB | Display | |
| Data in CIF |  emd_29737_validation.cif.gz emd_29737_validation.cif.gz | 26.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29737 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29737 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29737 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29737 | HTTPS FTP |
-Related structure data
| Related structure data |  8g5aMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_29737.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_29737.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 1 | ||||||||||||||||||||
| Voxel size | X=Y=Z: 0.825 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: X-31 Hemagglutinin full length soluble ectodomain trimer in...
| File | emd_29737_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | X-31 Hemagglutinin full length soluble ectodomain trimer in complex with three FL-1061 Fabs | ||||||||||||
| Projections & Slices |
| ||||||||||||
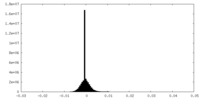
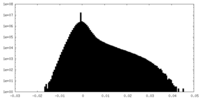
| Density Histograms |
-Half map: half map 2
| File | emd_29737_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
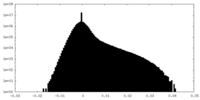
| Density Histograms |
- Sample components
Sample components
-Entire : X-31 hemagglutinin in complex with FL-1061 Fabs
| Entire | Name: X-31 hemagglutinin in complex with FL-1061 Fabs |
|---|---|
| Components |
|
-Supramolecule #1: X-31 hemagglutinin in complex with FL-1061 Fabs
| Supramolecule | Name: X-31 hemagglutinin in complex with FL-1061 Fabs / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:   Influenza A virus Influenza A virus |
-Macromolecule #1: Hemagglutinin
| Macromolecule | Name: Hemagglutinin / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Influenza A virus Influenza A virus |
| Molecular weight | Theoretical: 70.408609 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MKRGLCCVLL LCGAVFVSPS ASQDLPGNDN STATLCLGHH AVPNGTLVKT ITDDQIEVTN ATELVQSSST GKICNNPHRI LDGIDCTLI DALLGDPHCD VFQNETWDLF VERSKAFSNC YPYDVPDYAS LRSLVASSGT LEFITEGFTW TGVTQNGGSN A CKRGPGSG ...String: MKRGLCCVLL LCGAVFVSPS ASQDLPGNDN STATLCLGHH AVPNGTLVKT ITDDQIEVTN ATELVQSSST GKICNNPHRI LDGIDCTLI DALLGDPHCD VFQNETWDLF VERSKAFSNC YPYDVPDYAS LRSLVASSGT LEFITEGFTW TGVTQNGGSN A CKRGPGSG FFSRLNWLTK SGSTYPVLNV TMPNNDNFDK LYIWGIHHPS TDQEQTSLYV QASGRVTVST RRSQQTIIPN IG SRPWVRG LSSRISIYWT IVKPGDVLVI NSNGNLIAPR GYFKMRTGKS SIMRSDAPID TCISECITPN GSIPNDKPFQ NVN KITYGA CPKYVKQNTL KLATGMRNVP EKQTRGLFGA IAGFIENGWE GMIDGWYGFR HQNSEGTGQA ADLKSTQAAI DQIN GKLNR VIEKTNEKFH QIEKEFSEVE GRIQDLEKYV EDTKIDLWSY NAELLVALEN QHTIDLTDSE MNKLFEKTRR QLREN AEDM GNGCFKIYHK CDNACIESIR NGTYDHDVYR DEALNNRFQI KGVELKSGAG SSLEVLFQGP GSGSSLGGSG YIPEAP RDG QAYVRKDGEW VLLSTFLGSG SSHHHHHHHH GGSGSSMDEK TTGWRGGHVV EGLAGELEQL RARLEHHPQG QREP UniProtKB: Hemagglutinin |
-Macromolecule #2: FL-1086 Fab heavy chain
| Macromolecule | Name: FL-1086 Fab heavy chain / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 26.019004 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: ASQVQLQQSG AELMQPGASV KLSCKATGYT FAGYWIEWVK QRPGHGLEWI GEILPGIGST NYNGKFKGKA TFTADSSSNT AYMELSSLT TEDSAIYYCA RSGAQATFAM DYWGQGTSVT VSGASTKGPS VFPLAPSSKS TSGGTAALGC LVKDYFPEPV T VSWNSGAL ...String: ASQVQLQQSG AELMQPGASV KLSCKATGYT FAGYWIEWVK QRPGHGLEWI GEILPGIGST NYNGKFKGKA TFTADSSSNT AYMELSSLT TEDSAIYYCA RSGAQATFAM DYWGQGTSVT VSGASTKGPS VFPLAPSSKS TSGGTAALGC LVKDYFPEPV T VSWNSGAL TSGVHTFPAV LQSSGLYSLS SVVTVPSSSL GTQTYICNVN HKPSNTKVDK RVEPKSCDKG SSLEVLFQGP LG HHHHHH |
-Macromolecule #3: FL-1086 light chain
| Macromolecule | Name: FL-1086 light chain / type: protein_or_peptide / ID: 3 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 23.693311 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: ASDVQMTQSP SYLAASPGET ITINCRASKS ISKFLAWYQE KPGKTNKLLI YSGSTLQSGI PSRFSGSGSG TDFTLTISSL EPEDFAMYY CQQHNEYPYT FGAGTKLELK RTVAAPSVFI FPPSDEQLKS GTASVVCLLN NFYPREAKVQ WKVDNALQSG N SQESVTEQ ...String: ASDVQMTQSP SYLAASPGET ITINCRASKS ISKFLAWYQE KPGKTNKLLI YSGSTLQSGI PSRFSGSGSG TDFTLTISSL EPEDFAMYY CQQHNEYPYT FGAGTKLELK RTVAAPSVFI FPPSDEQLKS GTASVVCLLN NFYPREAKVQ WKVDNALQSG N SQESVTEQ DSKDSTYSLS STLTLSKADY EKHKVYACEV THQGLSSPVT KSFNRGEC |
-Macromolecule #4: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 4 / Number of copies: 12 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Grid | Model: C-flat-1.2/1.3 / Material: COPPER / Mesh: 400 / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Instrument: GATAN CRYOPLUNGE 3 |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 1.2 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.6 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller










 Z
Z Y
Y X
X