[English] 日本語
 Yorodumi
Yorodumi- EMDB-29645: Cryo-EM structure of an orphan GPCR-Gi protein signaling complex -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of an orphan GPCR-Gi protein signaling complex | ||||||||||||
 Map data Map data | EM map | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | GPCR / agonist / 6-OAU / MEMBRANE PROTEIN | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationurotensin II receptor activity / tertiary granule membrane / neuropeptide signaling pathway / specific granule membrane / Adenylate cyclase inhibitory pathway / positive regulation of protein localization to cell cortex / T cell migration / D2 dopamine receptor binding / response to prostaglandin E / G protein-coupled serotonin receptor binding ...urotensin II receptor activity / tertiary granule membrane / neuropeptide signaling pathway / specific granule membrane / Adenylate cyclase inhibitory pathway / positive regulation of protein localization to cell cortex / T cell migration / D2 dopamine receptor binding / response to prostaglandin E / G protein-coupled serotonin receptor binding / adenylate cyclase-inhibiting serotonin receptor signaling pathway / adenylate cyclase regulator activity / cellular response to forskolin / regulation of mitotic spindle organization / Regulation of insulin secretion / centriolar satellite / positive regulation of cholesterol biosynthetic process / G protein-coupled receptor binding / adenylate cyclase-inhibiting G protein-coupled receptor signaling pathway / adenylate cyclase-modulating G protein-coupled receptor signaling pathway / response to peptide hormone / G-protein beta/gamma-subunit complex binding / Olfactory Signaling Pathway / Activation of the phototransduction cascade / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Thromboxane signalling through TP receptor / G protein-coupled acetylcholine receptor signaling pathway / G-protein activation / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / Prostacyclin signalling through prostacyclin receptor / G beta:gamma signalling through CDC42 / Glucagon signaling in metabolic regulation / G beta:gamma signalling through BTK / Synthesis, secretion, and inactivation of Glucagon-like Peptide-1 (GLP-1) / ADP signalling through P2Y purinoceptor 12 / Sensory perception of sweet, bitter, and umami (glutamate) taste / photoreceptor disc membrane / Glucagon-type ligand receptors / Adrenaline,noradrenaline inhibits insulin secretion / Vasopressin regulates renal water homeostasis via Aquaporins / G alpha (z) signalling events / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / GDP binding / ADORA2B mediated anti-inflammatory cytokines production / cellular response to catecholamine stimulus / ADP signalling through P2Y purinoceptor 1 / G beta:gamma signalling through PI3Kgamma / adenylate cyclase-activating dopamine receptor signaling pathway / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / GPER1 signaling / Inactivation, recovery and regulation of the phototransduction cascade / cellular response to prostaglandin E stimulus / G-protein beta-subunit binding / heterotrimeric G-protein complex / G alpha (12/13) signalling events / sensory perception of taste / extracellular vesicle / signaling receptor complex adaptor activity / Thrombin signalling through proteinase activated receptors (PARs) / G protein activity / GTPase binding / Ca2+ pathway / retina development in camera-type eye / cell cortex / midbody / High laminar flow shear stress activates signaling by PIEZO1 and PECAM1:CDH5:KDR in endothelial cells / fibroblast proliferation / G alpha (i) signalling events / phospholipase C-activating G protein-coupled receptor signaling pathway / G alpha (s) signalling events / ciliary basal body / Hydrolases; Acting on acid anhydrides; Acting on GTP to facilitate cellular and subcellular movement / G alpha (q) signalling events / Ras protein signal transduction / Extra-nuclear estrogen signaling / cell population proliferation / receptor complex / G protein-coupled receptor signaling pathway / lysosomal membrane / cell division / GTPase activity / synapse / centrosome / Neutrophil degranulation / protein-containing complex binding / GTP binding / nucleolus / magnesium ion binding / Golgi apparatus / signal transduction / extracellular exosome / nucleoplasm / membrane / plasma membrane / cytosol / cytoplasm Similarity search - Function | ||||||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | ||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.0 Å | ||||||||||||
 Authors Authors | Zhang X / Wang YJ / Li X / Liu GB / Gong WM / Zhang C | ||||||||||||
| Funding support |  United States, United States,  China, 3 items China, 3 items
| ||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Pro-phagocytic function and structural basis of GPR84 signaling. Authors: Xuan Zhang / Yujing Wang / Shreyas Supekar / Xu Cao / Jingkai Zhou / Jessica Dang / Siqi Chen / Laura Jenkins / Sara Marsango / Xiu Li / Guibing Liu / Graeme Milligan / Mingye Feng / Hao Fan ...Authors: Xuan Zhang / Yujing Wang / Shreyas Supekar / Xu Cao / Jingkai Zhou / Jessica Dang / Siqi Chen / Laura Jenkins / Sara Marsango / Xiu Li / Guibing Liu / Graeme Milligan / Mingye Feng / Hao Fan / Weimin Gong / Cheng Zhang /     Abstract: GPR84 is a unique orphan G protein-coupled receptor (GPCR) that can be activated by endogenous medium-chain fatty acids (MCFAs). The signaling of GPR84 is largely pro-inflammatory, which can augment ...GPR84 is a unique orphan G protein-coupled receptor (GPCR) that can be activated by endogenous medium-chain fatty acids (MCFAs). The signaling of GPR84 is largely pro-inflammatory, which can augment inflammatory response, and GPR84 also functions as a pro-phagocytic receptor to enhance phagocytic activities of macrophages. In this study, we show that the activation of GPR84 by the synthetic agonist 6-OAU can synergize with the blockade of CD47 on cancer cells to induce phagocytosis of cancer cells by macrophages. We also determine a high-resolution structure of the GPR84-G signaling complex with 6-OAU. This structure reveals an occluded binding pocket for 6-OAU, the molecular basis of receptor activation involving non-conserved structural motifs of GPR84, and an unusual G-coupling interface. Together with computational docking and simulations studies, this structure also suggests a mechanism for the high selectivity of GPR84 for MCFAs and a potential routes of ligand binding and dissociation. These results provide a framework for understanding GPR84 signaling and developing new drugs targeting GPR84. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_29645.map.gz emd_29645.map.gz | 18.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-29645-v30.xml emd-29645-v30.xml emd-29645.xml emd-29645.xml | 20.5 KB 20.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_29645.png emd_29645.png | 500.5 KB | ||
| Filedesc metadata |  emd-29645.cif.gz emd-29645.cif.gz | 6.9 KB | ||
| Others |  emd_29645_half_map_1.map.gz emd_29645_half_map_1.map.gz emd_29645_half_map_2.map.gz emd_29645_half_map_2.map.gz | 20.7 MB 20.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-29645 http://ftp.pdbj.org/pub/emdb/structures/EMD-29645 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29645 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29645 | HTTPS FTP |
-Validation report
| Summary document |  emd_29645_validation.pdf.gz emd_29645_validation.pdf.gz | 825.2 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_29645_full_validation.pdf.gz emd_29645_full_validation.pdf.gz | 824.8 KB | Display | |
| Data in XML |  emd_29645_validation.xml.gz emd_29645_validation.xml.gz | 10.4 KB | Display | |
| Data in CIF |  emd_29645_validation.cif.gz emd_29645_validation.cif.gz | 12.1 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29645 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29645 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29645 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29645 | HTTPS FTP |
-Related structure data
| Related structure data |  8g05MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_29645.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_29645.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | EM map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.07 Å | ||||||||||||||||||||||||||||||||||||
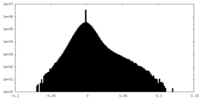
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: half1 map
| File | emd_29645_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half1 map | ||||||||||||
| Projections & Slices |
| ||||||||||||
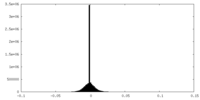
| Density Histograms |
-Half map: half2 map
| File | emd_29645_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half2 map | ||||||||||||
| Projections & Slices |
| ||||||||||||
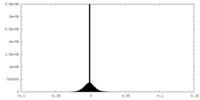
| Density Histograms |
- Sample components
Sample components
-Entire : 6OAU_GPR84_DNGi_scFv16
| Entire | Name: 6OAU_GPR84_DNGi_scFv16 |
|---|---|
| Components |
|
-Supramolecule #1: 6OAU_GPR84_DNGi_scFv16
| Supramolecule | Name: 6OAU_GPR84_DNGi_scFv16 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#5 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: G-protein coupled receptor 84
| Macromolecule | Name: G-protein coupled receptor 84 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 48.699938 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: DYKDDDDHHH HHHHHGQPGN GSAFLLAPNG SHAPDHNVTQ QRDEENLYFQ GVDMWNSSDA NFSCYHESVL GYRYVAVSWG VVVAVTGTV GNVLTLLALA IQPKLRTRFN LLIANLTLAD LLYCTLLQPF SVDTYLHLHW RTGATFCRVF GLLLFASNSV S ILTLCLIA ...String: DYKDDDDHHH HHHHHGQPGN GSAFLLAPNG SHAPDHNVTQ QRDEENLYFQ GVDMWNSSDA NFSCYHESVL GYRYVAVSWG VVVAVTGTV GNVLTLLALA IQPKLRTRFN LLIANLTLAD LLYCTLLQPF SVDTYLHLHW RTGATFCRVF GLLLFASNSV S ILTLCLIA LGRYLLIAHP KLFPQVFSAK GIVLALVSTW VVGVASFAPL WPIYILVPVV CTCSFDRIRG RPYTTILMGI YF VLGLSSV GIFYCLIHRQ VKRAAQALDQ YKLRQASIHS NHVARTDEAM PGRFQELDSR LASGGPSEGI SSEPVSAATT QTL EGDSSE VGDQINSKRA KQMAEKSPPE ASAKAQPIKG ARRAPDSSSE FGKVTRMCFA VFLCFALSYI PFLLLNILDA RVQA PRVVH MLAANLTWLN GCINPVLYAA MNRQFRQAYG SILKR UniProtKB: G-protein coupled receptor 84 |
-Macromolecule #2: Guanine nucleotide-binding protein G(i) subunit alpha-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(i) subunit alpha-1 type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 40.414047 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGCTLSAEDK AAVERSKMID RNLREDGEKA AREVKLLLLG AGESGKNTIV KQMKIIHEAG YSEEECKQYK AVVYSNTIQS IIAIIRAMG RLKIDFGDSA RADDARQLFV LAGAAEEGFM TAELAGVIKR LWKDSGVQAC FNRSREYQLN DSAAYYLNDL D RIAQPNYI ...String: MGCTLSAEDK AAVERSKMID RNLREDGEKA AREVKLLLLG AGESGKNTIV KQMKIIHEAG YSEEECKQYK AVVYSNTIQS IIAIIRAMG RLKIDFGDSA RADDARQLFV LAGAAEEGFM TAELAGVIKR LWKDSGVQAC FNRSREYQLN DSAAYYLNDL D RIAQPNYI PTQQDVLRTR VKTTGIVETH FTFKDLHFKM FDVGAQRSER KKWIHCFEGV TAIIFCVALS DYDLVLAEDE EM NRMHASM KLFDSICNNK WFTDTSIILF LNKKDLFEEK IKKSPLTICY PEYAGSNTYE EAAAYIQCQF EDLNKRKDTK EIY THFTCS TDTKNVQFVF DAVTDVIIKN NLKDCGLF UniProtKB: Guanine nucleotide-binding protein G(i) subunit alpha-1 |
-Macromolecule #3: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 40.845559 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MHHHHHHGSS GSELDQLRQE AEQLKNQIRD ARKACADATL SQITNNIDPV GRIQMRTRRT LRGHLAKIYA MHWGTDSRLL VSASQDGKL IIWDSYTTNK VHAIPLRSSW VMTCAYAPSG NYVACGGLDN ICSIYNLKTR EGNVRVSREL AGHTGYLSCC R FLDDNQIV ...String: MHHHHHHGSS GSELDQLRQE AEQLKNQIRD ARKACADATL SQITNNIDPV GRIQMRTRRT LRGHLAKIYA MHWGTDSRLL VSASQDGKL IIWDSYTTNK VHAIPLRSSW VMTCAYAPSG NYVACGGLDN ICSIYNLKTR EGNVRVSREL AGHTGYLSCC R FLDDNQIV TSSGDTTCAL WDIETGQQTT TFTGHTGDVM SLSLAPDTRL FVSGACDASA KLWDVREGMC RQTFTGHESD IN AICFFPN GNAFATGSDD ATCRLFDLRA DQELMTYSHD NIICGITSVS FSKSGRLLLA GYDDFNCNVW DALKADRAGV LAG HDNRVS CLGVTDDGMA VATGSWDSFL KIWNGSSGGG GSGGGGSSGV SGWRLFKKIS UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 |
-Macromolecule #4: scFv16
| Macromolecule | Name: scFv16 / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 28.694805 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: DVQLVESGGG LVQPGGSRKL SCSASGFAFS SFGMHWVRQA PEKGLEWVAY ISSGSGTIYY ADTVKGRFTI SRDDPKNTLF LQMTSLRSE DTAMYYCVRS IYYYGSSPFD FWGQGTTLTV SSGGGGSGGG GSGGGGSDIV MTQATSSVPV TPGESVSISC R SSKSLLHS ...String: DVQLVESGGG LVQPGGSRKL SCSASGFAFS SFGMHWVRQA PEKGLEWVAY ISSGSGTIYY ADTVKGRFTI SRDDPKNTLF LQMTSLRSE DTAMYYCVRS IYYYGSSPFD FWGQGTTLTV SSGGGGSGGG GSGGGGSDIV MTQATSSVPV TPGESVSISC R SSKSLLHS NGNTYLYWFL QRPGQSPQLL IYRMSNLASG VPDRFSGSGS GTAFTLTISR LEAEDVGVYY CMQHLEYPLT FG AGTKLEL LEENLYFQGA SHHHHHHHH |
-Macromolecule #5: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 7.861143 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MASNNTASIA QARKLVEQLK MEANIDRIKV SKAAADLMAY CEAHAKEDPL LTPVPASENP FREKKFFCAI L UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 |
-Macromolecule #6: 6-(octylamino)pyrimidine-2,4(3H,5H)-dione
| Macromolecule | Name: 6-(octylamino)pyrimidine-2,4(3H,5H)-dione / type: ligand / ID: 6 / Number of copies: 1 / Formula: YI9 |
|---|---|
| Molecular weight | Theoretical: 239.314 Da |
| Chemical component information | 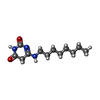 ChemComp-YI9: |
-Macromolecule #7: CHOLESTEROL
| Macromolecule | Name: CHOLESTEROL / type: ligand / ID: 7 / Number of copies: 1 / Formula: CLR |
|---|---|
| Molecular weight | Theoretical: 386.654 Da |
| Chemical component information |  ChemComp-CLR: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 55.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.2 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: EMDB MAP EMDB ID: |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.0 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 62864 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller






























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)