+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | LSD-bound serotonin 1A (5-HT1A) receptor-Gi1 protein complex | ||||||||||||||||||
 Map data Map data | LSD-bound serotonin 1A (5-HT1A) receptor-Gi1 protein complex | ||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||
 Keywords Keywords | GPCR Signaling Complex / Serotonin Receptor / SIGNALING PROTEIN | ||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationregulation of serotonin secretion / adenylate cyclase-inhibiting serotonin receptor signaling pathway / regulation of hormone secretion / regulation of behavior / Serotonin receptors / receptor-receptor interaction / serotonin receptor signaling pathway / regulation of dopamine metabolic process / serotonin metabolic process / serotonin binding ...regulation of serotonin secretion / adenylate cyclase-inhibiting serotonin receptor signaling pathway / regulation of hormone secretion / regulation of behavior / Serotonin receptors / receptor-receptor interaction / serotonin receptor signaling pathway / regulation of dopamine metabolic process / serotonin metabolic process / serotonin binding / G protein-coupled serotonin receptor activity / gamma-aminobutyric acid signaling pathway / exploration behavior / neurotransmitter receptor activity / G protein-coupled receptor signaling pathway, coupled to cyclic nucleotide second messenger / behavioral fear response / regulation of vasoconstriction / T cell migration / D2 dopamine receptor binding / Adenylate cyclase inhibitory pathway / positive regulation of protein localization to cell cortex / regulation of cAMP-mediated signaling / G protein-coupled serotonin receptor binding / cellular response to forskolin / regulation of mitotic spindle organization / adenylate cyclase-inhibiting G protein-coupled receptor signaling pathway / Regulation of insulin secretion / G protein-coupled receptor binding / G-protein beta/gamma-subunit complex binding / Olfactory Signaling Pathway / adenylate cyclase-modulating G protein-coupled receptor signaling pathway / Activation of the phototransduction cascade / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Thromboxane signalling through TP receptor / G protein-coupled acetylcholine receptor signaling pathway / G-protein activation / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / Prostacyclin signalling through prostacyclin receptor / response to peptide hormone / Glucagon signaling in metabolic regulation / G beta:gamma signalling through CDC42 / G beta:gamma signalling through BTK / ADP signalling through P2Y purinoceptor 12 / Sensory perception of sweet, bitter, and umami (glutamate) taste / Synthesis, secretion, and inactivation of Glucagon-like Peptide-1 (GLP-1) / photoreceptor disc membrane / Glucagon-type ligand receptors / Adrenaline,noradrenaline inhibits insulin secretion / Vasopressin regulates renal water homeostasis via Aquaporins / G alpha (z) signalling events / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / cellular response to catecholamine stimulus / ADORA2B mediated anti-inflammatory cytokines production / sensory perception of taste / ADP signalling through P2Y purinoceptor 1 / G beta:gamma signalling through PI3Kgamma / adenylate cyclase-activating dopamine receptor signaling pathway / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / GPER1 signaling / GDP binding / cellular response to prostaglandin E stimulus / Inactivation, recovery and regulation of the phototransduction cascade / G-protein beta-subunit binding / heterotrimeric G-protein complex / G alpha (12/13) signalling events / extracellular vesicle / signaling receptor complex adaptor activity / Thrombin signalling through proteinase activated receptors (PARs) / GTPase binding / retina development in camera-type eye / Ca2+ pathway / cell cortex / phospholipase C-activating G protein-coupled receptor signaling pathway / midbody / G alpha (i) signalling events / fibroblast proliferation / G alpha (s) signalling events / Hydrolases; Acting on acid anhydrides; Acting on GTP to facilitate cellular and subcellular movement / chemical synaptic transmission / G alpha (q) signalling events / Ras protein signal transduction / cell population proliferation / Extra-nuclear estrogen signaling / G protein-coupled receptor signaling pathway / lysosomal membrane / cell division / GTPase activity / centrosome / positive regulation of cell population proliferation / dendrite / synapse / protein-containing complex binding / GTP binding / nucleolus / magnesium ion binding / signal transduction / extracellular exosome / nucleoplasm Similarity search - Function | ||||||||||||||||||
| Biological species | Escherichia coli, Homo sapiens /  Homo sapiens (human) Homo sapiens (human) | ||||||||||||||||||
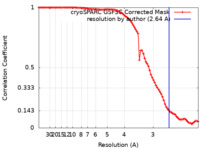
| Method | single particle reconstruction / cryo EM / Resolution: 2.64 Å | ||||||||||||||||||
 Authors Authors | Warren AL / Zilberg G / Capper MJ / Wacker D | ||||||||||||||||||
| Funding support |  United States, 5 items United States, 5 items
| ||||||||||||||||||
 Citation Citation |  Journal: Nature / Year: 2024 Journal: Nature / Year: 2024Title: Structural pharmacology and therapeutic potential of 5-methoxytryptamines. Authors: Audrey L Warren / David Lankri / Michael J Cunningham / Inis C Serrano / Lyonna F Parise / Andrew C Kruegel / Priscilla Duggan / Gregory Zilberg / Michael J Capper / Vaclav Havel / Scott J ...Authors: Audrey L Warren / David Lankri / Michael J Cunningham / Inis C Serrano / Lyonna F Parise / Andrew C Kruegel / Priscilla Duggan / Gregory Zilberg / Michael J Capper / Vaclav Havel / Scott J Russo / Dalibor Sames / Daniel Wacker /  Abstract: Psychedelic substances such as lysergic acid diethylamide (LSD) and psilocybin show potential for the treatment of various neuropsychiatric disorders. These compounds are thought to mediate their ...Psychedelic substances such as lysergic acid diethylamide (LSD) and psilocybin show potential for the treatment of various neuropsychiatric disorders. These compounds are thought to mediate their hallucinogenic and therapeutic effects through the serotonin (5-hydroxytryptamine (5-HT)) receptor 5-HT (ref. ). However, 5-HT also plays a part in the behavioural effects of tryptamine hallucinogens, particularly 5-methoxy-N,N-dimethyltryptamine (5-MeO-DMT), a psychedelic found in the toxin of Colorado River toads. Although 5-HT is a validated therapeutic target, little is known about how psychedelics engage 5-HT and which effects are mediated by this receptor. Here we map the molecular underpinnings of 5-MeO-DMT pharmacology through five cryogenic electron microscopy (cryo-EM) structures of 5-HT, systematic medicinal chemistry, receptor mutagenesis and mouse behaviour. Structure-activity relationship analyses of 5-methoxytryptamines at both 5-HT and 5-HT enable the characterization of molecular determinants of 5-HT signalling potency, efficacy and selectivity. Moreover, we contrast the structural interactions and in vitro pharmacology of 5-MeO-DMT and analogues to the pan-serotonergic agonist LSD and clinically used 5-HT agonists. We show that a 5-HT-selective 5-MeO-DMT analogue is devoid of hallucinogenic-like effects while retaining anxiolytic-like and antidepressant-like activity in socially defeated animals. Our studies uncover molecular aspects of 5-HT-targeted psychedelics and therapeutics, which may facilitate the future development of new medications for neuropsychiatric disorders. | ||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_29597.map.gz emd_29597.map.gz | 31.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-29597-v30.xml emd-29597-v30.xml emd-29597.xml emd-29597.xml | 24.3 KB 24.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_29597_fsc.xml emd_29597_fsc.xml | 8.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_29597.png emd_29597.png | 36 KB | ||
| Filedesc metadata |  emd-29597.cif.gz emd-29597.cif.gz | 7.9 KB | ||
| Others |  emd_29597_half_map_1.map.gz emd_29597_half_map_1.map.gz emd_29597_half_map_2.map.gz emd_29597_half_map_2.map.gz | 59.4 MB 59.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-29597 http://ftp.pdbj.org/pub/emdb/structures/EMD-29597 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29597 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29597 | HTTPS FTP |
-Validation report
| Summary document |  emd_29597_validation.pdf.gz emd_29597_validation.pdf.gz | 843.6 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_29597_full_validation.pdf.gz emd_29597_full_validation.pdf.gz | 843.2 KB | Display | |
| Data in XML |  emd_29597_validation.xml.gz emd_29597_validation.xml.gz | 16.6 KB | Display | |
| Data in CIF |  emd_29597_validation.cif.gz emd_29597_validation.cif.gz | 21.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29597 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29597 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29597 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29597 | HTTPS FTP |
-Related structure data
| Related structure data |  8fytMC  8fy8C  8fyeC  8fylC  8fyxC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_29597.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_29597.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | LSD-bound serotonin 1A (5-HT1A) receptor-Gi1 protein complex | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.058 Å | ||||||||||||||||||||||||||||||||||||
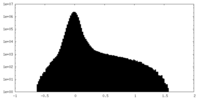
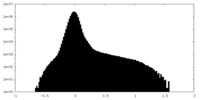
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Half Map 1
| File | emd_29597_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
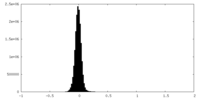
| Density Histograms |
-Half map: Half Map 2
| File | emd_29597_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
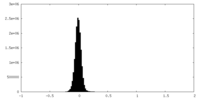
| Density Histograms |
- Sample components
Sample components
-Entire : Serotonin 1A (5-HT1A) receptor-Gi1 protein complex
| Entire | Name: Serotonin 1A (5-HT1A) receptor-Gi1 protein complex |
|---|---|
| Components |
|
-Supramolecule #1: Serotonin 1A (5-HT1A) receptor-Gi1 protein complex
| Supramolecule | Name: Serotonin 1A (5-HT1A) receptor-Gi1 protein complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 Details: Serotonin 1A (5-HT1A) receptor-Gi1 protein complex with components purified separately and assembled in vitro. |
|---|---|
| Source (natural) | Organism: Escherichia coli, Homo sapiens |
-Macromolecule #1: Soluble cytochrome b562,5-hydroxytryptamine receptor 1A,5-hydroxy...
| Macromolecule | Name: Soluble cytochrome b562,5-hydroxytryptamine receptor 1A,5-hydroxytryptamine receptor 1A type: protein_or_peptide / ID: 1 Details: N-terminal HA signal sequence, flag-tag, his-tag, tev cleavage site followed by soluble cytochrome b562 (E. coli) and truncated 5-hydroxytryptamine receptor 1A (Homo sapiens) with a point ...Details: N-terminal HA signal sequence, flag-tag, his-tag, tev cleavage site followed by soluble cytochrome b562 (E. coli) and truncated 5-hydroxytryptamine receptor 1A (Homo sapiens) with a point mutation at position 255 in the provided sequence. This mutation is in Ballesteros-Weinstein position 3.41 of 5-HT1A where the original leucine was mutated to a tryptophan. Cytochrome b562 (cybC, UniProt P0ABE7) is both n and c-terminally truncated with two point mutations.,N-terminal HA signal sequence, flag-tag, his-tag, tev cleavage site followed by soluble cytochrome b562 (E. coli) and truncated 5-hydroxytryptamine receptor 1A (Homo sapiens) with a point mutation at position 255 in the provided sequence. This mutation is in Ballesteros-Weinstein position 3.41 of 5-HT1A where the original leucine was mutated to a tryptophan. Cytochrome b562 (cybC, UniProt P0ABE7) is both n and c-terminally truncated with two point mutations. Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 61.37593 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MKTIIALSYI FCLVFADYKD DDDAKLQTMH HHHHHHHHHE NLYFQGGTTA DLEDNWETLN DNLKVIEKAD NAAQVKDALT KMRAAALDA QKATPPKLED KSPDSPEMKD FRHGFDILVG QIDDALKLAN EGKVKEAQAA AEQLKTTRNA YIQKYLTGIS D VTVSYQVI ...String: MKTIIALSYI FCLVFADYKD DDDAKLQTMH HHHHHHHHHE NLYFQGGTTA DLEDNWETLN DNLKVIEKAD NAAQVKDALT KMRAAALDA QKATPPKLED KSPDSPEMKD FRHGFDILVG QIDDALKLAN EGKVKEAQAA AEQLKTTRNA YIQKYLTGIS D VTVSYQVI TSLLLGTLIF CAVLGNACVV AAIALERSLQ NVANYLIGSL AVTDLMVSVL VLPMAALYQV LNKWTLGQVT CD LFIALDV LCCTSSIWHL CAIALDRYWA ITDPIDYVNK RTPRRAAALI SLTWLIGFLI SIPPMLGWRT PEDRSDPDAC TIS KDHGYT IYSTFGAFYI PLLLMLVLYG RIFRAARFRI RKTVKKVEKT GADTRHGASP APQPKKSVNG ESGSRNWRLG VESK AGGAL CANGAVRQGD DGAALEVIEV HRVGNSKEHL PLPSEAGPTP CAPASFERKN ERNAEAKRKM ALARERKTVK TLGII MGTF ILCWLPFFIV ALVLPFCESS CHMPTLLGAI INWLGYSNSL LNPVIYAYFN KDFQNAFKKI IKCKFCRQ UniProtKB: 5-hydroxytryptamine receptor 1A |
-Macromolecule #2: Guanine nucleotide-binding protein G(i) subunit alpha-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(i) subunit alpha-1 type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 40.415031 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGCTLSAEDK AAVERSKMID RNLREDGEKA AREVKLLLLG AGESGKSTIV KQMKIIHEAG YSEEECKQYK AVVYSNTIQS IIAIIRAMG RLKIDFGDSA RADDARQLFV LAGAAEEGFM TAELAGVIKR LWKDSGVQAC FNRSREYQLN DSAAYYLNDL D RIAQPNYI ...String: MGCTLSAEDK AAVERSKMID RNLREDGEKA AREVKLLLLG AGESGKSTIV KQMKIIHEAG YSEEECKQYK AVVYSNTIQS IIAIIRAMG RLKIDFGDSA RADDARQLFV LAGAAEEGFM TAELAGVIKR LWKDSGVQAC FNRSREYQLN DSAAYYLNDL D RIAQPNYI PTQQDVLRTR VKTTGIVETH FTFKDLHFKM FDVGGQRSER KKWIHCFEGV TAIIFCVALS DYDLVLAEDE EM NRMHESM KLFDSICNNK WFTDTSIILF LNKKDLFEEK IKKSPLTICY PEYAGSNTYE EAAAYIQCQF EDLNKRKDTK EIY THFTCA TDTKNVQFVF DAVTDVIIKN NLKDCGLF UniProtKB: Guanine nucleotide-binding protein G(i) subunit alpha-1 |
-Macromolecule #3: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 type: protein_or_peptide / ID: 3 Details: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 with N-terminal his-tag and 3C cleavage site and GSSG linker Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 39.418086 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MHHHHHHLEV LFQGPGSSGS ELDQLRQEAE QLKNQIRDAR KACADATLSQ ITNNIDPVGR IQMRTRRTLR GHLAKIYAMH WGTDSRLLV SASQDGKLII WDSYTTNKVH AIPLRSSWVM TCAYAPSGNY VACGGLDNIC SIYNLKTREG NVRVSRELAG H TGYLSCCR ...String: MHHHHHHLEV LFQGPGSSGS ELDQLRQEAE QLKNQIRDAR KACADATLSQ ITNNIDPVGR IQMRTRRTLR GHLAKIYAMH WGTDSRLLV SASQDGKLII WDSYTTNKVH AIPLRSSWVM TCAYAPSGNY VACGGLDNIC SIYNLKTREG NVRVSRELAG H TGYLSCCR FLDDNQIVTS SGDTTCALWD IETGQQTTTF TGHTGDVMSL SLAPDTRLFV SGACDASAKL WDVREGMCRQ TF TGHESDI NAICFFPNGN AFATGSDDAT CRLFDLRADQ ELMTYSHDNI ICGITSVSFS KSGRLLLAGY DDFNCNVWDA LKA DRAGVL AGHDNRVSCL GVTDDGMAVA TGSWDSFLKI WN UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 |
-Macromolecule #4: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 8.506765 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MASNNTASIA QARKLVEQLK MEANIDRIKV SKAAADLMAY CEAHAKEDPL LTPVPASENP FREKKFFCAI LGSAGSAGSA UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 |
-Macromolecule #5: (8alpha)-N,N-diethyl-6-methyl-9,10-didehydroergoline-8-carboxamide
| Macromolecule | Name: (8alpha)-N,N-diethyl-6-methyl-9,10-didehydroergoline-8-carboxamide type: ligand / ID: 5 / Number of copies: 1 / Formula: 7LD |
|---|---|
| Molecular weight | Theoretical: 323.432 Da |
| Chemical component information |  ChemComp-7LD: |
-Macromolecule #6: [(2R)-1-[oxidanyl-[(2R,3R,5S,6R)-2,3,5,6-tetrakis(oxidanyl)-4-pho...
| Macromolecule | Name: [(2R)-1-[oxidanyl-[(2R,3R,5S,6R)-2,3,5,6-tetrakis(oxidanyl)-4-phosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-3-tetradecanoyloxy-propan-2-yl] (5E,8E)-hexadeca-5,8,11,14-tetraenoate type: ligand / ID: 6 / Number of copies: 1 / Formula: J40 |
|---|---|
| Molecular weight | Theoretical: 854.895 Da |
| Chemical component information | 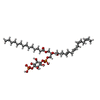 ChemComp-J40: |
-Macromolecule #7: CHOLESTEROL HEMISUCCINATE
| Macromolecule | Name: CHOLESTEROL HEMISUCCINATE / type: ligand / ID: 7 / Number of copies: 2 / Formula: Y01 |
|---|---|
| Molecular weight | Theoretical: 486.726 Da |
| Chemical component information |  ChemComp-Y01: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 26 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
| ||||||||||||
| Grid | Model: Quantifoil / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Instrument: LEICA EM GP Details: Blot force 3 for 3-5 seconds was used and subsequent grids were screened for ice thickness prior to data collection.. | ||||||||||||
| Details | Sample was mono disperse following gel filtration. The sample was immediately concentrated for CryoEM grid preparation. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 53.61 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller




































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)