+ データを開く
データを開く
- 基本情報
基本情報
| 登録情報 |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| タイトル | Cryo-EM structure of Stanieria sp. CphA2 | |||||||||
 マップデータ マップデータ | Stanieria sp. CphA2 locally filtered map | |||||||||
 試料 試料 |
| |||||||||
 キーワード キーワード | cyanophycin / CphA2 / ligase / ATP-grasp | |||||||||
| 機能・相同性 |  機能・相同性情報 機能・相同性情報ribosomal S6-glutamic acid ligase activity / SOS response / ATP binding / metal ion binding / cytoplasm 類似検索 - 分子機能 | |||||||||
| 生物種 |  Stanieria sp. (バクテリア) / Stanieria sp. (バクテリア) /  Stanieria sp. NIES-3757 (バクテリア) Stanieria sp. NIES-3757 (バクテリア) | |||||||||
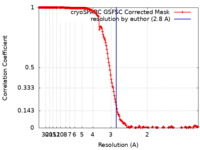
| 手法 | 単粒子再構成法 / クライオ電子顕微鏡法 / 解像度: 2.8 Å | |||||||||
 データ登録者 データ登録者 | Markus LM / Sharon I / Strauss M / Schmeing TM | |||||||||
| 資金援助 |  カナダ, 1件 カナダ, 1件
| |||||||||
 引用 引用 |  ジャーナル: Protein Sci / 年: 2023 ジャーナル: Protein Sci / 年: 2023タイトル: Structure and function of a hexameric cyanophycin synthetase 2. 著者: Linda M D Markus / Itai Sharon / Kim Munro / Marcel Grogg / Donald Hilvert / Mike Strauss / T Martin Schmeing /   要旨: Cyanophycin is a natural polymer composed of a poly-aspartate backbone with arginine attached to each of the aspartate sidechains. Produced by a wide range of bacteria, which mainly use it as a store ...Cyanophycin is a natural polymer composed of a poly-aspartate backbone with arginine attached to each of the aspartate sidechains. Produced by a wide range of bacteria, which mainly use it as a store of fixed nitrogen, it has many promising industrial applications. Cyanophycin can be synthesized from the amino acids Asp and Arg by the widespread cyanophycin synthetase 1 (CphA1), or from the dipeptide β-Asp-Arg by the cyanobacterial enzyme cyanophycin synthetase 2 (CphA2). CphA2 enzymes display a range of oligomeric states, from dimers to dodecamers. Recently, the crystal structure of a CphA2 dimer was solved but could not be obtained in complex with substrate. Here, we report cryo-EM structures of the hexameric CphA2 from Stanieria sp. at ~2.8 Å resolution, both with and without ATP analog and cyanophycin. The structures show a two-fold symmetrical, trimer-of-dimers hexameric architecture, and substrate-binding interactions that are similar to those of CphA1. Mutagenesis experiments demonstrate the importance of several conserved substrate-binding residues. We also find that a Q416A/R528G double mutation prevents hexamer formation and use this double mutant to show that hexamerization augments the rate of cyanophycin synthesis. Together, these results increase our mechanistic understanding of how an interesting green polymer is biosynthesized. | |||||||||
| 履歴 |
|
- 構造の表示
構造の表示
| 添付画像 |
|---|
- ダウンロードとリンク
ダウンロードとリンク
-EMDBアーカイブ
| マップデータ |  emd_29533.map.gz emd_29533.map.gz | 22.4 MB |  EMDBマップデータ形式 EMDBマップデータ形式 | |
|---|---|---|---|---|
| ヘッダ (付随情報) |  emd-29533-v30.xml emd-29533-v30.xml emd-29533.xml emd-29533.xml | 14 KB 14 KB | 表示 表示 |  EMDBヘッダ EMDBヘッダ |
| FSC (解像度算出) |  emd_29533_fsc.xml emd_29533_fsc.xml | 17 KB | 表示 |  FSCデータファイル FSCデータファイル |
| 画像 |  emd_29533.png emd_29533.png | 64 KB | ||
| マスクデータ |  emd_29533_msk_1.map emd_29533_msk_1.map | 512 MB |  マスクマップ マスクマップ | |
| Filedesc metadata |  emd-29533.cif.gz emd-29533.cif.gz | 5.5 KB | ||
| その他 |  emd_29533_half_map_1.map.gz emd_29533_half_map_1.map.gz emd_29533_half_map_2.map.gz emd_29533_half_map_2.map.gz | 474.2 MB 474.2 MB | ||
| アーカイブディレクトリ |  http://ftp.pdbj.org/pub/emdb/structures/EMD-29533 http://ftp.pdbj.org/pub/emdb/structures/EMD-29533 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29533 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29533 | HTTPS FTP |
-検証レポート
| 文書・要旨 |  emd_29533_validation.pdf.gz emd_29533_validation.pdf.gz | 1 MB | 表示 |  EMDB検証レポート EMDB検証レポート |
|---|---|---|---|---|
| 文書・詳細版 |  emd_29533_full_validation.pdf.gz emd_29533_full_validation.pdf.gz | 1 MB | 表示 | |
| XML形式データ |  emd_29533_validation.xml.gz emd_29533_validation.xml.gz | 26.6 KB | 表示 | |
| CIF形式データ |  emd_29533_validation.cif.gz emd_29533_validation.cif.gz | 34.5 KB | 表示 | |
| アーカイブディレクトリ |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29533 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29533 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29533 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29533 | HTTPS FTP |
-関連構造データ
| 関連構造データ |  8fxhMC  8fxiC M: このマップから作成された原子モデル C: 同じ文献を引用 ( |
|---|---|
| 類似構造データ | 類似検索 - 機能・相同性  F&H 検索 F&H 検索 |
- リンク
リンク
| EMDBのページ |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| 「今月の分子」の関連する項目 |
- マップ
マップ
| ファイル |  ダウンロード / ファイル: emd_29533.map.gz / 形式: CCP4 / 大きさ: 512 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) ダウンロード / ファイル: emd_29533.map.gz / 形式: CCP4 / 大きさ: 512 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | Stanieria sp. CphA2 locally filtered map | ||||||||||||||||||||||||||||||||||||
| 投影像・断面図 | 画像のコントロール
画像は Spider により作成 | ||||||||||||||||||||||||||||||||||||
| ボクセルのサイズ | X=Y=Z: 0.675 Å | ||||||||||||||||||||||||||||||||||||
| 密度 |
| ||||||||||||||||||||||||||||||||||||
| 対称性 | 空間群: 1 | ||||||||||||||||||||||||||||||||||||
| 詳細 | EMDB XML:
|
-添付データ
-マスク #1
| ファイル |  emd_29533_msk_1.map emd_29533_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 投影像・断面図 |
| ||||||||||||
| 密度ヒストグラム |
-ハーフマップ: Stanieria sp. CphA2 half map A
| ファイル | emd_29533_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | Stanieria sp. CphA2 half map A | ||||||||||||
| 投影像・断面図 |
| ||||||||||||
| 密度ヒストグラム |
-ハーフマップ: Stanieria sp. CphA2 half map B
| ファイル | emd_29533_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | Stanieria sp. CphA2 half map B | ||||||||||||
| 投影像・断面図 |
| ||||||||||||
| 密度ヒストグラム |
- 試料の構成要素
試料の構成要素
-全体 : Stanieria sp. CphA2
| 全体 | 名称: Stanieria sp. CphA2 |
|---|---|
| 要素 |
|
-超分子 #1: Stanieria sp. CphA2
| 超分子 | 名称: Stanieria sp. CphA2 / タイプ: complex / ID: 1 / 親要素: 0 / 含まれる分子: all |
|---|---|
| 由来(天然) | 生物種:  Stanieria sp. (バクテリア) Stanieria sp. (バクテリア) |
-分子 #1: RimK domain-containing protein ATP-grasp
| 分子 | 名称: RimK domain-containing protein ATP-grasp / タイプ: protein_or_peptide / ID: 1 / コピー数: 6 / 光学異性体: LEVO |
|---|---|
| 由来(天然) | 生物種:  Stanieria sp. NIES-3757 (バクテリア) Stanieria sp. NIES-3757 (バクテリア) |
| 分子量 | 理論値: 72.886516 KDa |
| 組換発現 | 生物種:  |
| 配列 | 文字列: MLTKQAVEPV RINARTTDVF DIFNVKQYVG ANPYLNQAAL VFDFAFTESY QPLPIENYLA VVGDRYPRLK EIEYQSYAEL FASTVAEVN KLEMDLHLKG WNVKPIEEIN RIAIESLHHR TTKEVVYCVW DWFEFITQGE EFDLSKQIAI LQQLFRNSVY G GPTVYALL ...文字列: MLTKQAVEPV RINARTTDVF DIFNVKQYVG ANPYLNQAAL VFDFAFTESY QPLPIENYLA VVGDRYPRLK EIEYQSYAEL FASTVAEVN KLEMDLHLKG WNVKPIEEIN RIAIESLHHR TTKEVVYCVW DWFEFITQGE EFDLSKQIAI LQQLFRNSVY G GPTVYALL RTANEKHIPA FYLWDEGLMQ YGYGKQQVRG IATTFDVDSH IDSDFTTQKD DCKKFLQELG FPVPQGDVVF SL AEAKEVA AEIGYPVAVK PVAGHKGIGV TADVQDEIEL EAAYDRAVAG IPLEEKICII VENSIAGHDY RLLCVNGRFV AAT ERKPAY VVGDGYSTIA ELIEKENFSP NRSDTPTSPM GKIRTDEAMH LYLEEQGLDL DSVIDRDRTI YLRKVANLSS GGFS IDATN RVHPDNIILA QDIAQHFRLT CLGIDIITND IGRSWKETSF GIIEINAAPG VYMHLKPAIG EPVDVTARIL ETFFE TEKN ARIPIITFNR VSIRQLQKLS DRILMSHPDW TIGAVCREGI LINRSEKILN RHYNTNVLNL LRNPKLDLLI AEYDED ALE AEGMFYHGSN LVVLEDPSEI EMILTRDVFS DSTVIIKQGR EITIKRKGLL EQYELEAEEL IEQVYLKEIG TISENLY FQ UniProtKB: RimK domain-containing protein ATP-grasp |
-実験情報
-構造解析
| 手法 | クライオ電子顕微鏡法 |
|---|---|
 解析 解析 | 単粒子再構成法 |
| 試料の集合状態 | particle |
- 試料調製
試料調製
| 緩衝液 | pH: 7.5 |
|---|---|
| 凍結 | 凍結剤: ETHANE |
- 電子顕微鏡法
電子顕微鏡法
| 顕微鏡 | FEI TITAN KRIOS |
|---|---|
| 撮影 | フィルム・検出器のモデル: GATAN K3 BIOQUANTUM (6k x 4k) 平均電子線量: 80.0 e/Å2 |
| 電子線 | 加速電圧: 300 kV / 電子線源:  FIELD EMISSION GUN FIELD EMISSION GUN |
| 電子光学系 | 照射モード: SPOT SCAN / 撮影モード: BRIGHT FIELD / 最大 デフォーカス(公称値): -0.0025 µm / 最小 デフォーカス(公称値): -0.001 µm |
| 実験機器 |  モデル: Titan Krios / 画像提供: FEI Company |
 ムービー
ムービー コントローラー
コントローラー









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)