+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Neck structure of Agrobacterium phage Milano, C3 symmetry | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Myophage / redox trigger / VIRUS | |||||||||
| Function / homology |  Function and homology information Function and homology information | |||||||||
| Biological species |  Agrobacterium phage Milano (virus) Agrobacterium phage Milano (virus) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.46 Å | |||||||||
 Authors Authors | Sonani RR / Wang F / Esteves NC / Kelly RJ / Sebastian A / Kreutzberger MAB / Leiman PG / Scharf BE / Egelman EH | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Commun Biol / Year: 2023 Journal: Commun Biol / Year: 2023Title: Neck and capsid architecture of the robust Agrobacterium phage Milano. Authors: Ravi R Sonani / Nathaniel C Esteves / Abigail A Horton / Rebecca J Kelly / Amanda L Sebastian / Fengbin Wang / Mark A B Kreutzberger / Petr G Leiman / Birgit E Scharf / Edward H Egelman /  Abstract: Large gaps exist in our understanding of how bacteriophages, the most abundant biological entities on Earth, assemble and function. The structure of the "neck" region, where the DNA-filled capsid is ...Large gaps exist in our understanding of how bacteriophages, the most abundant biological entities on Earth, assemble and function. The structure of the "neck" region, where the DNA-filled capsid is connected to the host-recognizing tail remains poorly understood. We describe cryo-EM structures of the neck, the neck-capsid and neck-tail junctions, and capsid of the Agrobacterium phage Milano. The Milano neck 1 protein connects the 12-fold symmetrical neck to a 5-fold vertex of the icosahedral capsid. Comparison of Milano neck 1 homologs leads to four proposed classes, likely evolved from the simplest one in siphophages to more complex ones in myo- and podophages. Milano neck is surrounded by the atypical collar, which covalently crosslinks the tail sheath to neck 1. The Milano capsid is decorated with three types of proteins, a minor capsid protein (mCP) and two linking proteins crosslinking the mCP to the major capsid protein. The extensive network of disulfide bonds within and between neck, collar, capsid and tail provides an exceptional structural stability to Milano. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_29503.map.gz emd_29503.map.gz | 322.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-29503-v30.xml emd-29503-v30.xml emd-29503.xml emd-29503.xml | 18.9 KB 18.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_29503_fsc.xml emd_29503_fsc.xml | 14.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_29503.png emd_29503.png | 129.9 KB | ||
| Filedesc metadata |  emd-29503.cif.gz emd-29503.cif.gz | 6.1 KB | ||
| Others |  emd_29503_half_map_1.map.gz emd_29503_half_map_1.map.gz emd_29503_half_map_2.map.gz emd_29503_half_map_2.map.gz | 317.9 MB 317.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-29503 http://ftp.pdbj.org/pub/emdb/structures/EMD-29503 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29503 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29503 | HTTPS FTP |
-Validation report
| Summary document |  emd_29503_validation.pdf.gz emd_29503_validation.pdf.gz | 1.4 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_29503_full_validation.pdf.gz emd_29503_full_validation.pdf.gz | 1.4 MB | Display | |
| Data in XML |  emd_29503_validation.xml.gz emd_29503_validation.xml.gz | 24.1 KB | Display | |
| Data in CIF |  emd_29503_validation.cif.gz emd_29503_validation.cif.gz | 31.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29503 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29503 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29503 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29503 | HTTPS FTP |
-Related structure data
| Related structure data |  8fweMC  8fwbC  8fwcC  8fwgC  8fwmC  8fxpC  8fxrC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_29503.map.gz / Format: CCP4 / Size: 343 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_29503.map.gz / Format: CCP4 / Size: 343 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.08 Å | ||||||||||||||||||||||||||||||||||||
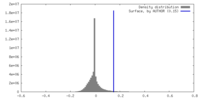
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_29503_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
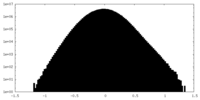
| Density Histograms |
-Half map: #2
| File | emd_29503_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Agrobacterium phage Milano
| Entire | Name:  Agrobacterium phage Milano (virus) Agrobacterium phage Milano (virus) |
|---|---|
| Components |
|
-Supramolecule #1: Agrobacterium phage Milano
| Supramolecule | Name: Agrobacterium phage Milano / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all / NCBI-ID: 2557550 / Sci species name: Agrobacterium phage Milano / Virus type: VIRION / Virus isolate: SPECIES / Virus enveloped: Yes / Virus empty: No |
|---|---|
| Host (natural) | Organism:  Agrobacterium fabrum str. C58 (bacteria) Agrobacterium fabrum str. C58 (bacteria) |
-Macromolecule #1: Collar sheath protein, gp13
| Macromolecule | Name: Collar sheath protein, gp13 / type: protein_or_peptide / ID: 1 / Number of copies: 60 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Agrobacterium phage Milano (virus) Agrobacterium phage Milano (virus) |
| Molecular weight | Theoretical: 24.490402 KDa |
| Sequence | String: MYFFSVDPRN GASKSGDVCG SCCCESISAR PGEVNGVMVS YAAWSAPLRG HGLTNKTTFE IDGVSVTPPK VSNAFGRTKV GVVFEGTLS DLFPNPEGEQ VEYEISELNG PSNGVVELGA NGAFTYTPGA LFTGVDRFWF SINGNIGEYV ISVDPTTSEL P QPPFTTPV ...String: MYFFSVDPRN GASKSGDVCG SCCCESISAR PGEVNGVMVS YAAWSAPLRG HGLTNKTTFE IDGVSVTPPK VSNAFGRTKV GVVFEGTLS DLFPNPEGEQ VEYEISELNG PSNGVVELGA NGAFTYTPGA LFTGVDRFWF SINGNIGEYV ISVDPTTSEL P QPPFTTPV YVPAARRSVD PRTHVLKFVL GVSPAAIPGD VYRLTVRQVA IDCDGNEFVH ISCYDISIGS CG UniProtKB: Virion-associated protein |
-Macromolecule #2: Portal protein, gp7
| Macromolecule | Name: Portal protein, gp7 / type: protein_or_peptide / ID: 2 / Number of copies: 12 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Agrobacterium phage Milano (virus) Agrobacterium phage Milano (virus) |
| Molecular weight | Theoretical: 45.840844 KDa |
| Sequence | String: MLGIPLLTRK AALTPPAPSA NPAKIFIRRF FSAGVAKNVV SYSNVMAAQR AMEHPVAFRC LDKLGLTVQS VKWDVGKDPQ NTQVGDGGM SASQRKALQQ ILQRPNPTMS GAQLRYSAAL SWACFGRMAF KVSVMSDGSV NAIWPLGIPF LKQKFDRYGD V ESFQYGDE ...String: MLGIPLLTRK AALTPPAPSA NPAKIFIRRF FSAGVAKNVV SYSNVMAAQR AMEHPVAFRC LDKLGLTVQS VKWDVGKDPQ NTQVGDGGM SASQRKALQQ ILQRPNPTMS GAQLRYSAAL SWACFGRMAF KVSVMSDGSV NAIWPLGIPF LKQKFDRYGD V ESFQYGDE AGKETIPSFT KVEKNDKGRP IKNYAFMIVK PSINGAMNFD VQNTPLQAIG VPVALYDALM ARAIDSADGT PN SKWLVTA SRDLDDGQAK EVKEGIEETK PGGDNGGEII FIAGTDVKVQ EMKNDLSDIH SKVPLDDQAR TIAGNFGIPI ALL GFAGAD GSKFANNYDE SRKAFFEDTI EPGYLTPLED GFSMFLCGAG YRVIFDRDSI PALRKSRADI AATYDKVTFI TEEE KREVT GWPAKKEGQT QNDDA UniProtKB: Portal protein |
-Macromolecule #3: Neck 2 protein, gp15
| Macromolecule | Name: Neck 2 protein, gp15 / type: protein_or_peptide / ID: 3 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Agrobacterium phage Milano (virus) Agrobacterium phage Milano (virus) |
| Molecular weight | Theoretical: 16.052353 KDa |
| Sequence | String: MRTADRKHRV IVCSQQSDVD DEGRLLITRA GVIQGWAAIA PVKAIRFSQD GVSMQKDTMQ PTHDITMNYN PDVNVSVSAW VYEHRLKSP PRWFKVLSVV NVDECSRYMK IRCRLVETSD DVTPPVEEEK NSFGAVKIDI PL UniProtKB: Head completion protein |
-Macromolecule #4: Tail-terminator protein, gp18
| Macromolecule | Name: Tail-terminator protein, gp18 / type: protein_or_peptide / ID: 4 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Agrobacterium phage Milano (virus) Agrobacterium phage Milano (virus) |
| Molecular weight | Theoretical: 20.268541 KDa |
| Sequence | String: METKLTYGNR VTLPEFAKYI VAPAFHEIEG RAIPVTGVDD DASGTQATKL PFVLVGLRQG DTSGPATIAG NSTINLRDDF IVEFNMKKE RYRDRKGGET PFFSYYDYES IRDRLFNSMI EFSGEHGITF EFVSLDISTE GDVVYIEFRF RQNYEWCETV R EADTTIEA GRFSINLQGC UniProtKB: Virion-associated protein |
-Macromolecule #5: Tail-tube, gp21
| Macromolecule | Name: Tail-tube, gp21 / type: protein_or_peptide / ID: 5 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Agrobacterium phage Milano (virus) Agrobacterium phage Milano (virus) |
| Molecular weight | Theoretical: 14.673427 KDa |
| Sequence | String: MACNKQNGVK NILITFTDCD TQEVIGPISH EQPDDTLPTY KNCAWTNTAL TNGYVQRSAS NATMTLPVVR DLRVPLAFYQ GCAQVDVQV EKFDGTVMTL TEGAVVEPEE SDGRSVTMNI VASEIDELLP PGSLAAA UniProtKB: Virion-associated protein |
-Macromolecule #6: Neck 1 protein, gp14
| Macromolecule | Name: Neck 1 protein, gp14 / type: protein_or_peptide / ID: 6 / Number of copies: 12 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Agrobacterium phage Milano (virus) Agrobacterium phage Milano (virus) |
| Molecular weight | Theoretical: 22.255439 KDa |
| Sequence | String: MNLDTLLPLQ TIREHAKCDD NPRVTDDLLK LYREAAFEAA ELYTGLSFTP EKTIVEPIRL KGRRGKIILS ATPIAGRPVV FYGGGLGSP LELIPRPGSN VLFFPYGSPD RFQTWGDCHT CDVESQLMAT YVTGRRCENS VPAGIIIGIL KLIAWNINNP G DEVMSVRN ...String: MNLDTLLPLQ TIREHAKCDD NPRVTDDLLK LYREAAFEAA ELYTGLSFTP EKTIVEPIRL KGRRGKIILS ATPIAGRPVV FYGGGLGSP LELIPRPGSN VLFFPYGSPD RFQTWGDCHT CDVESQLMAT YVTGRRCENS VPAGIIIGIL KLIAWNINNP G DEVMSVRN TLNANAQGLI GGTNNGAVIS GAQDEWFRYR RVLL UniProtKB: Head-to-tail connector complex protein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.2 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)