+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | 40S subunit of the Giardia lamblia 80S ribosome | ||||||||||||
 Map data Map data | Main map for building the body of the 40S subunit. | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | Giardia lamblia / Ribosome structure / Translation / parasite / RIBOSOME | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationendonucleolytic cleavage to generate mature 3'-end of SSU-rRNA from (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / maturation of SSU-rRNA / maintenance of translational fidelity / ribosomal small subunit biogenesis / small ribosomal subunit rRNA binding / ribosomal small subunit assembly / small ribosomal subunit / cytosolic small ribosomal subunit ...endonucleolytic cleavage to generate mature 3'-end of SSU-rRNA from (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / maturation of SSU-rRNA / maintenance of translational fidelity / ribosomal small subunit biogenesis / small ribosomal subunit rRNA binding / ribosomal small subunit assembly / small ribosomal subunit / cytosolic small ribosomal subunit / rRNA binding / ribosome / structural constituent of ribosome / ribonucleoprotein complex / translation / mRNA binding / nucleolus / RNA binding / zinc ion binding / nucleus / cytosol Similarity search - Function | ||||||||||||
| Biological species |  Giardia intestinalis assemblage A (eukaryote) Giardia intestinalis assemblage A (eukaryote) | ||||||||||||
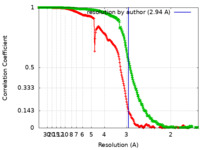
| Method | single particle reconstruction / cryo EM / Resolution: 2.94 Å | ||||||||||||
 Authors Authors | Eiler DR / Wimberly BT / Bilodeau DY / Rissland OS / Kieft JS | ||||||||||||
| Funding support |  United States, 3 items United States, 3 items
| ||||||||||||
 Citation Citation |  Journal: Structure / Year: 2024 Journal: Structure / Year: 2024Title: The Giardia lamblia ribosome structure reveals divergence in several biological pathways and the mode of emetine function. Authors: Daniel R Eiler / Brian T Wimberly / Danielle Y Bilodeau / J Matthew Taliaferro / Philip Reigan / Olivia S Rissland / Jeffrey S Kieft /  Abstract: Giardia lamblia is a deeply branching protist and a human pathogen. Its unusual biology presents the opportunity to explore conserved and fundamental molecular mechanisms. We determined the structure ...Giardia lamblia is a deeply branching protist and a human pathogen. Its unusual biology presents the opportunity to explore conserved and fundamental molecular mechanisms. We determined the structure of the G. lamblia 80S ribosome bound to tRNA, mRNA, and the antibiotic emetine by cryo-electron microscopy, to an overall resolution of 2.49 Å. The structure reveals rapidly evolving protein and nucleotide regions, differences in the peptide exit tunnel, and likely altered ribosome quality control pathways. Examination of translation initiation factor binding sites suggests these interactions are conserved despite a divergent initiation mechanism. Highlighting the potential of G. lamblia to resolve conserved biological principles; our structure reveals the interactions of the translation inhibitor emetine with the ribosome and mRNA, thus providing insight into the mechanism of action for this widely used antibiotic. Our work defines key questions in G. lamblia and motivates future experiments to explore the diversity of eukaryotic gene regulation. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_29495.map.gz emd_29495.map.gz | 321 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-29495-v30.xml emd-29495-v30.xml emd-29495.xml emd-29495.xml | 54 KB 54 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_29495_fsc.xml emd_29495_fsc.xml emd_29495_fsc_2.xml emd_29495_fsc_2.xml | 24.3 KB 24.3 KB | Display Display |  FSC data file FSC data file |
| Images |  emd_29495.png emd_29495.png | 45.4 KB | ||
| Filedesc metadata |  emd-29495.cif.gz emd-29495.cif.gz | 11 KB | ||
| Others |  emd_29495_additional_1.map.gz emd_29495_additional_1.map.gz emd_29495_additional_2.map.gz emd_29495_additional_2.map.gz emd_29495_half_map_1.map.gz emd_29495_half_map_1.map.gz emd_29495_half_map_2.map.gz emd_29495_half_map_2.map.gz | 314.7 MB 321.1 MB 1.2 GB 1.2 GB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-29495 http://ftp.pdbj.org/pub/emdb/structures/EMD-29495 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29495 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29495 | HTTPS FTP |
-Validation report
| Summary document |  emd_29495_validation.pdf.gz emd_29495_validation.pdf.gz | 1.3 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_29495_full_validation.pdf.gz emd_29495_full_validation.pdf.gz | 1.3 MB | Display | |
| Data in XML |  emd_29495_validation.xml.gz emd_29495_validation.xml.gz | 33 KB | Display | |
| Data in CIF |  emd_29495_validation.cif.gz emd_29495_validation.cif.gz | 42.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29495 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29495 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29495 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29495 | HTTPS FTP |
-Related structure data
| Related structure data |  8fvyMC  8fruC  8g4iC  8g4sC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_29495.map.gz / Format: CCP4 / Size: 347.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_29495.map.gz / Format: CCP4 / Size: 347.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Main map for building the body of the 40S subunit. | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.8211 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: Main map for building the head of the 40S subunit.
| File | emd_29495_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Main map for building the head of the 40S subunit. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Main map for refinement of the 40S subunit.
| File | emd_29495_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Main map for refinement of the 40S subunit. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map of focused refinement P191 J466 Focused Head 2p94.mrc
| File | emd_29495_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map of focused refinement P191_J466_Focused_Head_2p94.mrc | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map of focused refinement P191 J466 Focused Head 2p94.mrc
| File | emd_29495_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map of focused refinement P191_J466_Focused_Head_2p94.mrc | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
+Entire : 60S ribosomal subunit of the 80S Giardia intestinalis assemblage ...
+Supramolecule #1: 60S ribosomal subunit of the 80S Giardia intestinalis assemblage ...
+Macromolecule #1: 18S rRNA
+Macromolecule #32: tRNA
+Macromolecule #2: 40S ribosomal protein SA
+Macromolecule #3: 40S ribosomal protein S3a
+Macromolecule #4: Ribosomal protein S2
+Macromolecule #5: Ribosomal protein S3
+Macromolecule #6: 40S ribosomal protein S4
+Macromolecule #7: SSU ribosomal protein S7P (Fragment)
+Macromolecule #8: 40S ribosomal protein S6
+Macromolecule #9: 40S ribosomal protein S7
+Macromolecule #10: 40S ribosomal protein S8
+Macromolecule #11: Ribosomal protein S9
+Macromolecule #12: Ribosomal protein S10B
+Macromolecule #13: SSU ribosomal protein S17P
+Macromolecule #14: Ribosomal protein S13
+Macromolecule #15: Ribosomal protein S14
+Macromolecule #16: Ribosomal protein S15
+Macromolecule #17: Ribosomal protein S16
+Macromolecule #18: Ribosomal protein S17
+Macromolecule #19: Ribosomal protein S18
+Macromolecule #20: SSU ribosomal protein S19E (Fragment)
+Macromolecule #21: Ribosomal protein S20
+Macromolecule #22: 40S ribosomal protein S21
+Macromolecule #23: SSU ribosomal protein S8P (Fragment)
+Macromolecule #24: SSU ribosomal protein S12P
+Macromolecule #25: Ribosomal protein S24
+Macromolecule #26: 40S ribosomal protein S25
+Macromolecule #27: 40S ribosomal protein S26
+Macromolecule #28: Ribosomal protein S27
+Macromolecule #29: Ribosomal protein S28
+Macromolecule #30: Ribosomal protein S29A
+Macromolecule #31: Ribosomal protein S27a
+Macromolecule #33: Ribosomal protein S12
+Macromolecule #34: 40S ribosomal protein S30
+Macromolecule #35: MAGNESIUM ION
+Macromolecule #36: emetine
+Macromolecule #37: ZINC ION
+Macromolecule #38: water
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 72.26 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.9000000000000001 µm / Nominal defocus min: 0.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller




















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)