[English] 日本語
 Yorodumi
Yorodumi- EMDB-29421: Full-length mouse 5-HT3A receptor in complex with SMP100, open-like -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Full-length mouse 5-HT3A receptor in complex with SMP100, open-like | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Partial agonist / cys-loop / pLGIC / ion channel / TRANSPORT PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationNeurotransmitter receptors and postsynaptic signal transmission / serotonin-gated monoatomic cation channel activity / serotonin-gated cation-selective signaling pathway / serotonin-activated cation-selective channel complex / serotonin receptor signaling pathway / serotonin binding / inorganic cation transmembrane transport / cleavage furrow / ligand-gated monoatomic ion channel activity involved in regulation of presynaptic membrane potential / postsynaptic membrane / identical protein binding Similarity search - Function | |||||||||
| Biological species |  | |||||||||
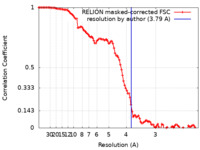
| Method | single particle reconstruction / cryo EM / Resolution: 3.79 Å | |||||||||
 Authors Authors | Felt KC / Chakrapani S | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2024 Journal: Nat Struct Mol Biol / Year: 2024Title: Structural basis for partial agonism in 5-HT receptors. Authors: Kevin Felt / Madeleine Stauffer / Leslie Salas-Estrada / Peter R Guzzo / Dejian Xie / Jinkun Huang / Marta Filizola / Sudha Chakrapani /   Abstract: Hyperactivity of serotonin 3 receptors (5-HTR) underlies pathologies associated with irritable bowel syndrome and chemotherapy-induced nausea and vomiting. Setrons, a class of high-affinity ...Hyperactivity of serotonin 3 receptors (5-HTR) underlies pathologies associated with irritable bowel syndrome and chemotherapy-induced nausea and vomiting. Setrons, a class of high-affinity competitive antagonists, are used in the treatment of these conditions. Although generally effective for chemotherapy-induced nausea and vomiting, the use of setrons for treating irritable bowel syndrome has been impaired by adverse side effects. Partial agonists are now being considered as an alternative strategy, with potentially less severe side effects than full antagonists. However, a structural understanding of how these ligands work is lacking. Here, we present high-resolution cryogenic electron microscopy structures of the mouse 5-HTR in complex with partial agonists (SMP-100 and ALB-148471) captured in pre-activated and open-like conformational states. Molecular dynamics simulations were used to assess the stability of drug-binding poses and interactions with the receptor over time. Together, these studies reveal mechanisms for the functional differences between orthosteric partial agonists, full agonists and antagonists of the 5-HTR. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_29421.map.gz emd_29421.map.gz | 96.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-29421-v30.xml emd-29421-v30.xml emd-29421.xml emd-29421.xml | 18.2 KB 18.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_29421_fsc.xml emd_29421_fsc.xml | 10.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_29421.png emd_29421.png | 52.8 KB | ||
| Filedesc metadata |  emd-29421.cif.gz emd-29421.cif.gz | 6.2 KB | ||
| Others |  emd_29421_half_map_1.map.gz emd_29421_half_map_1.map.gz emd_29421_half_map_2.map.gz emd_29421_half_map_2.map.gz | 80.8 MB 80.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-29421 http://ftp.pdbj.org/pub/emdb/structures/EMD-29421 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29421 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29421 | HTTPS FTP |
-Validation report
| Summary document |  emd_29421_validation.pdf.gz emd_29421_validation.pdf.gz | 972.5 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_29421_full_validation.pdf.gz emd_29421_full_validation.pdf.gz | 972.1 KB | Display | |
| Data in XML |  emd_29421_validation.xml.gz emd_29421_validation.xml.gz | 17.2 KB | Display | |
| Data in CIF |  emd_29421_validation.cif.gz emd_29421_validation.cif.gz | 22.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29421 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29421 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29421 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29421 | HTTPS FTP |
-Related structure data
| Related structure data |  8fspMC  8frwC  8frxC  8frzC  8fsbC  8fszC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_29421.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_29421.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.1 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_29421_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_29421_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : 5-HT3A pentamer in complex with orthosteric ligand SMP100
| Entire | Name: 5-HT3A pentamer in complex with orthosteric ligand SMP100 |
|---|---|
| Components |
|
-Supramolecule #1: 5-HT3A pentamer in complex with orthosteric ligand SMP100
| Supramolecule | Name: 5-HT3A pentamer in complex with orthosteric ligand SMP100 type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 331 KDa |
-Macromolecule #1: 5-hydroxytryptamine receptor 3A
| Macromolecule | Name: 5-hydroxytryptamine receptor 3A / type: protein_or_peptide / ID: 1 / Number of copies: 5 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 62.349812 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: WSHPQFEKGG GSGGGSGGGS WSHPQFEKGG GSGGGSGGGS WSHPQFEKGG GSGGGSGGGS WSHPQFEKEN LYFQGATQAR DTTQPALLR LSDHLLANYK KGVRPVRDWR KPTTVSIDVI MYAILNVDEK NQVLTTYIWY RQYWTDEFLQ WTPEDFDNVT K LSIPTDSI ...String: WSHPQFEKGG GSGGGSGGGS WSHPQFEKGG GSGGGSGGGS WSHPQFEKGG GSGGGSGGGS WSHPQFEKEN LYFQGATQAR DTTQPALLR LSDHLLANYK KGVRPVRDWR KPTTVSIDVI MYAILNVDEK NQVLTTYIWY RQYWTDEFLQ WTPEDFDNVT K LSIPTDSI WVPDILINEF VDVGKSPNIP YVYVHHRGEV QNYKPLQLVT ACSLDIYNFP FDVQNCSLTF TSWLHTIQDI NI TLWRSPE EVRSDKSIFI NQGEWELLEV FPQFKEFSID ISNSYAEMKF YVIIRRRPLF YAVSLLLPSI FLMVVDIVGF CLP PDSGER VSFKITLLLG YSVFLIIVSD TLPATAIGTP LIGVYFVVCM ALLVISLAET IFIVRLVHKQ DLQRPVPDWL RHLV LDRIA WILCLGEQPM AHRPPATFQA NKTDDCSGSD LLPAMGNHCS HVGGPQDLEK TPRGRGSPLP PPREASLAVR GLLQE LSSI RHFLEKRDEM REVARDWLRV GYVLDRLLFR IYLLAVLAYS ITLVTLWSIW HYSENLYFQG TETSQVAPA UniProtKB: 5-hydroxytryptamine receptor 3A |
-Macromolecule #2: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 2 / Number of copies: 15 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Macromolecule #3: 5-[(1R,3S,4R)-1-azabicyclo[2.2.2]octan-3-yl]-1,3,4,5-tetrahydro-6...
| Macromolecule | Name: 5-[(1R,3S,4R)-1-azabicyclo[2.2.2]octan-3-yl]-1,3,4,5-tetrahydro-6H-azepino[5,4,3-cd]indazol-6-one type: ligand / ID: 3 / Number of copies: 5 / Formula: Y82 |
|---|---|
| Molecular weight | Theoretical: 296.367 Da |
| Chemical component information | 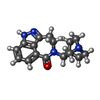 ChemComp-Y82: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2.6 mg/mL |
|---|---|
| Buffer | pH: 8 |
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Support film - Material: CARBON / Support film - topology: HOLEY |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK I |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 1.8 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 81000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL |
|---|---|
| Output model |  PDB-8fsp: |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)