[English] 日本語
 Yorodumi
Yorodumi- EMDB-29296: Cryo-EM structure of NLR family apoptosis inhibitory protein 5 (N... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of NLR family apoptosis inhibitory protein 5 (NAIP5) in complex with a full-length flagellin (FliC) ligand | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Inflammasome / Innate immunity / Bacterial ligand / host-pathogen interaction / Protein complex / IMMUNE SYSTEM | |||||||||
| Function / homology |  Function and homology information Function and homology informationTLR5 cascade / MyD88 cascade initiated on plasma membrane / NFkB and MAPK activation mediated by TRAF6 / IPAF inflammasome complex / The IPAF inflammasome / bacterial-type flagellum / pyroptotic inflammatory response / cysteine-type endopeptidase inhibitor activity involved in apoptotic process / detection of bacterium / positive regulation of interleukin-1 beta production ...TLR5 cascade / MyD88 cascade initiated on plasma membrane / NFkB and MAPK activation mediated by TRAF6 / IPAF inflammasome complex / The IPAF inflammasome / bacterial-type flagellum / pyroptotic inflammatory response / cysteine-type endopeptidase inhibitor activity involved in apoptotic process / detection of bacterium / positive regulation of interleukin-1 beta production / perikaryon / defense response to Gram-negative bacterium / receptor ligand activity / defense response to bacterium / inflammatory response / symbiont entry into host cell / innate immune response / neuronal cell body / negative regulation of apoptotic process / apoptotic process / structural molecule activity / extracellular space / ATP binding / metal ion binding Similarity search - Function | |||||||||
| Biological species |   Salmonella enterica subsp. enterica serovar Typhimurium str. LT2 (bacteria) Salmonella enterica subsp. enterica serovar Typhimurium str. LT2 (bacteria) | |||||||||
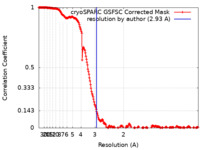
| Method | single particle reconstruction / cryo EM / Resolution: 2.93 Å | |||||||||
 Authors Authors | Paidimuddala B / Zhang L | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Sci Adv / Year: 2023 Journal: Sci Adv / Year: 2023Title: Structural basis for flagellin-induced NAIP5 activation. Authors: Bhaskar Paidimuddala / Jianhao Cao / Liman Zhang /  Abstract: The NAIP (NLR family apoptosis inhibitory protein)/NLRC4 (NLR family CARD containing protein 4) inflammasome senses Gram-negative bacterial ligand. In the ligand-bound state, the winged helix domain ...The NAIP (NLR family apoptosis inhibitory protein)/NLRC4 (NLR family CARD containing protein 4) inflammasome senses Gram-negative bacterial ligand. In the ligand-bound state, the winged helix domain of NAIP forms a steric clash with NLRC4 to open it up. However, how ligand binding activates NAIP is less clear. Here, we investigated the dynamics of the ligand-binding region of inactive NAIP5 and solved the cryo-EM structure of NAIP5 in complex with its specific ligand, FliC from flagellin, at 2.9-Å resolution. The structure revealed a "trap and lock" mechanism in FliC recognition, whereby FliC-D0 is first trapped by the hydrophobic pocket of NAIP5, then locked in the binding site by ID (insertion domain) and C-terminal tail of NAIP5. The FliC-D0 domain further inserts into ID to stabilize the complex. According to this mechanism, FliC triggers the conformational change of NAIP5 by bringing multiple flexible domains together. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_29296.map.gz emd_29296.map.gz | 785.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-29296-v30.xml emd-29296-v30.xml emd-29296.xml emd-29296.xml | 17.6 KB 17.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_29296_fsc.xml emd_29296_fsc.xml | 20.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_29296.png emd_29296.png | 120.6 KB | ||
| Filedesc metadata |  emd-29296.cif.gz emd-29296.cif.gz | 6.9 KB | ||
| Others |  emd_29296_half_map_1.map.gz emd_29296_half_map_1.map.gz emd_29296_half_map_2.map.gz emd_29296_half_map_2.map.gz | 877.2 MB 877.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-29296 http://ftp.pdbj.org/pub/emdb/structures/EMD-29296 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29296 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29296 | HTTPS FTP |
-Validation report
| Summary document |  emd_29296_validation.pdf.gz emd_29296_validation.pdf.gz | 1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_29296_full_validation.pdf.gz emd_29296_full_validation.pdf.gz | 1 MB | Display | |
| Data in XML |  emd_29296_validation.xml.gz emd_29296_validation.xml.gz | 29.7 KB | Display | |
| Data in CIF |  emd_29296_validation.cif.gz emd_29296_validation.cif.gz | 38.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29296 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29296 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29296 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29296 | HTTPS FTP |
-Related structure data
| Related structure data |  8fmlMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_29296.map.gz / Format: CCP4 / Size: 944.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_29296.map.gz / Format: CCP4 / Size: 944.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.528 Å | ||||||||||||||||||||||||||||||||||||
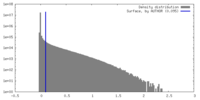
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_29296_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
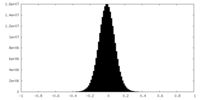
| Density Histograms |
-Half map: #1
| File | emd_29296_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
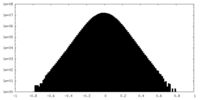
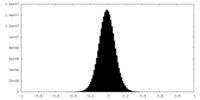
| Density Histograms |
- Sample components
Sample components
-Entire : NAIP5 in complex with a full-length bacterial flagellin ligand (FliC)
| Entire | Name: NAIP5 in complex with a full-length bacterial flagellin ligand (FliC) |
|---|---|
| Components |
|
-Supramolecule #1: NAIP5 in complex with a full-length bacterial flagellin ligand (FliC)
| Supramolecule | Name: NAIP5 in complex with a full-length bacterial flagellin ligand (FliC) type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 213.5 KDa |
-Macromolecule #1: Baculoviral IAP repeat-containing protein 1e
| Macromolecule | Name: Baculoviral IAP repeat-containing protein 1e / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 161.128156 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MDYKDDDDKL AEHGESSEDR ISEIDYEFLP ELSALLGVDA FQVAKSQEEE EHKERMKMKK GFNSQMRSEA KRLKTFETYD TFRSWTPQE MAAAGFYHTG VRLGVQCFCC SLILFGNSLR KLPIERHKKL RPECEFLQGK DVGNIGKYDI RVKRPEKMLR G GKARYHEE ...String: MDYKDDDDKL AEHGESSEDR ISEIDYEFLP ELSALLGVDA FQVAKSQEEE EHKERMKMKK GFNSQMRSEA KRLKTFETYD TFRSWTPQE MAAAGFYHTG VRLGVQCFCC SLILFGNSLR KLPIERHKKL RPECEFLQGK DVGNIGKYDI RVKRPEKMLR G GKARYHEE EARLESFEDW PFYAHGTSPR VLSAAGFVFT GKRDTVQCFS CGGSLGNWEE GDDPWKEHAK WFPKCEFLQS KK SSEEIAQ YIQSYEGFVH VTGEHFVKSW VRRELPMVSA YCNDSVFANE ELRMDMFKDW PQESPVGVEA LVRAGFFYTG KKD IVRCFS CGGCLEKWAE GDDPMEDHIK FFPECVFLQT LKSSAEVIPT LQSQYALPEA TETTRESNHG DAAAVHSTVV DLGR SEAQW FQEARSLSEQ LRDNYTKATF RHMNLPEVCS SLGTDHLLSC DVSIISKHIS QPVQEALTIP EVFSNLNSVM CVEGE TGSG KTTFLKRIAF LWASGCCPLL YRFQLVFYLS LSSITPDQGL ANIICAQLLG AGGCISEVCL SSSIQQLQHQ VLFLLD DYS GLASLPQALH TLITKNYLSR TCLLIAVHTN RVRDIRLYLG TSLEIQEFPF YNTVSVLRKF FSHDIICVEK LIIYFID NK DLQGVYKTPL FVAAVCTDWI QNASAQDKFQ DVTLFQSYMQ YLSLKYKATA EPLQATVSSC GQLALTGLFS SCFEFNSD D LAEAGVDEDE KLTTLLMSKF TAQRLRPVYR FLGPLFQEFL AAVRLTELLS SDRQEDQDLG LYYLRQIDSP LKAINSFNI FLYYVSSHSS SKAAPTVVSH LLQLVDEKES LENMSENEDY MKLHPQTFLW FQFVRGLWLV SPESSSSFVS EHLLRLALIF AYESNTVAE CSPFILQFLR GKTLALRVLN LQYFRDHPES LLLLRSLKVS INGNKMSSYV DYSFKTYFEN LQPPAIDEEY T SAFEHISE WRRNFAQDEE IIKNYENIRP RALPDISEGY WKLSPKPCKI PKLEVQVNNT DAADQALLQV LMEVFSASQS IE FRLFNSS GFLESICPAL ELSKASVTKC SMSRLELSRA EQELLLTLPA LQSLEVSETN QLPEQLFHNL HKFLGLKELC VRL DGKPNV LSVLPREFPN LLHMEKLSIQ TSTESDLSKL VKFIQNFPNL HVFHLKCDFL SNCESLMAVL ASCKKLREIE FSGR CFEAM TFVNILPNFV SLKILNLKDQ QFPDKETSEK FAQALGSLRN LEELLVPTGD GIHQVAKLIV RQCLQLPCLR VLTFH DILD DDSVIEIARA ATSGGFQKLE NLDISMNHKI TEEGYRNFFQ ALDNLPNLQE LNICRNIPGR IQVQATTVKA LGQCVS RLP SLIRLHMLSW LLDEEDMKVI NDVKERHPQS KRLIIFWKLI VPFSPVILE UniProtKB: Baculoviral IAP repeat-containing protein 1e |
-Macromolecule #2: Flagellin
| Macromolecule | Name: Flagellin / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Salmonella enterica subsp. enterica serovar Typhimurium str. LT2 (bacteria) Salmonella enterica subsp. enterica serovar Typhimurium str. LT2 (bacteria) |
| Molecular weight | Theoretical: 52.614418 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MHHHHHHMAQ VINTNSLSLL TQNNLNKSQS ALGTAIERLS SGLRINSAKD DAAGQAIANR FTANIKGLTQ ASRNANDGIS IAQTTEGAL NEINNNLQRV RELAVQSANS TNSQSDLDSI QAEITQRLNE IDRVSGQTQF NGVKVLAQDN TLTIQVGAND G ETIDIDLK ...String: MHHHHHHMAQ VINTNSLSLL TQNNLNKSQS ALGTAIERLS SGLRINSAKD DAAGQAIANR FTANIKGLTQ ASRNANDGIS IAQTTEGAL NEINNNLQRV RELAVQSANS TNSQSDLDSI QAEITQRLNE IDRVSGQTQF NGVKVLAQDN TLTIQVGAND G ETIDIDLK QINSQTLGLD TLNVQQKYKV SDTAATVTGY ADTTIALDNS TFKASATGLG GTDQKIDGDL KFDDTTGKYY AK VTVTGGT GKDGYYEVSV DKTNGEVTLA GGATSPLTGG LPATATEDVK NVQVANADLT EAKAALTAAG VTGTASVVKM SYT DNNGKT IDGGLAVKVG DDYYSATQNK DGSISINTTK YTADDGTSKT ALNKLGGADG KTEVVSIGGK TYAASKAEGH NFKA QPDLA EAAATTTENP LQKIDAALAQ VDTLRSDLGA VQNRFNSAIT NLGNTVNNLT SARSRIEDSD YATEVSNMSR AQILQ QAGT SVLAQANQVP QNVLSLLR UniProtKB: Flagellin |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | cell |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Grid | Model: UltrAuFoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Number grids imaged: 1 / Number real images: 4616 / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 81000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: BACKBONE TRACE |
|---|---|
| Output model |  PDB-8fml: |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)