+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Focused refinement of corner area2 for MapSPARTA | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | pAgo / SPARTA / Argonuate / Oligomerization / TIR domain / NAD / Immune system | |||||||||
| Biological species |  Maribacter polysiphoniae (bacteria) Maribacter polysiphoniae (bacteria) | |||||||||
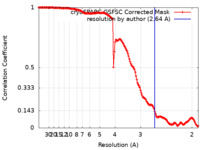
| Method | single particle reconstruction / cryo EM / Resolution: 2.64 Å | |||||||||
 Authors Authors | Shen ZF / Fu TM | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2023 Journal: Nature / Year: 2023Title: Oligomerization-mediated activation of a short prokaryotic Argonaute. Authors: Zhangfei Shen / Xiao-Yuan Yang / Shiyu Xia / Wei Huang / Derek J Taylor / Kotaro Nakanishi / Tian-Min Fu /  Abstract: Although eukaryotic and long prokaryotic Argonaute proteins (pAgos) cleave nucleic acids, some short pAgos lack nuclease activity and hydrolyse NAD(P) to induce bacterial cell death. Here we present ...Although eukaryotic and long prokaryotic Argonaute proteins (pAgos) cleave nucleic acids, some short pAgos lack nuclease activity and hydrolyse NAD(P) to induce bacterial cell death. Here we present a hierarchical activation pathway for SPARTA, a short pAgo consisting of an Argonaute (Ago) protein and TIR-APAZ, an associated protein. SPARTA progresses through distinct oligomeric forms, including a monomeric apo state, a monomeric RNA-DNA-bound state, two dimeric RNA-DNA-bound states and a tetrameric RNA-DNA-bound active state. These snapshots together identify oligomerization as a mechanistic principle of SPARTA activation. The RNA-DNA-binding channel of apo inactive SPARTA is occupied by an auto-inhibitory motif in TIR-APAZ. After the binding of RNA-DNA, SPARTA transitions from a monomer to a symmetric dimer and then an asymmetric dimer, in which two TIR domains interact through charge and shape complementarity. Next, two dimers assemble into a tetramer with a central TIR cluster responsible for hydrolysing NAD(P). In addition, we observe unique features of interactions between SPARTA and RNA-DNA, including competition between the DNA 3' end and the auto-inhibitory motif, interactions between the RNA G2 nucleotide and Ago, and splaying of the RNA-DNA duplex by two loops exclusive to short pAgos. Together, our findings provide a mechanistic basis for the activation of short pAgos, a large section of the Ago superfamily. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_29225.map.gz emd_29225.map.gz | 1.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-29225-v30.xml emd-29225-v30.xml emd-29225.xml emd-29225.xml | 11.9 KB 11.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_29225_fsc.xml emd_29225_fsc.xml | 14.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_29225.png emd_29225.png | 61.3 KB | ||
| Others |  emd_29225_half_map_1.map.gz emd_29225_half_map_1.map.gz emd_29225_half_map_2.map.gz emd_29225_half_map_2.map.gz | 285.4 MB 285.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-29225 http://ftp.pdbj.org/pub/emdb/structures/EMD-29225 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29225 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29225 | HTTPS FTP |
-Validation report
| Summary document |  emd_29225_validation.pdf.gz emd_29225_validation.pdf.gz | 747.4 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_29225_full_validation.pdf.gz emd_29225_full_validation.pdf.gz | 747 KB | Display | |
| Data in XML |  emd_29225_validation.xml.gz emd_29225_validation.xml.gz | 23.6 KB | Display | |
| Data in CIF |  emd_29225_validation.cif.gz emd_29225_validation.cif.gz | 30.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29225 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29225 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29225 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29225 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_29225.map.gz / Format: CCP4 / Size: 307.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_29225.map.gz / Format: CCP4 / Size: 307.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.95 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_29225_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_29225_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Structure of tetramerized short MapSPARTA (short prokarytic Argon...
| Entire | Name: Structure of tetramerized short MapSPARTA (short prokarytic Argonuate) system upon guide RNA-mediated target ssDNA binding |
|---|---|
| Components |
|
-Supramolecule #1: Structure of tetramerized short MapSPARTA (short prokarytic Argon...
| Supramolecule | Name: Structure of tetramerized short MapSPARTA (short prokarytic Argonuate) system upon guide RNA-mediated target ssDNA binding type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  Maribacter polysiphoniae (bacteria) Maribacter polysiphoniae (bacteria) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: DIRECT ELECTRON APOLLO (4k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.1 µm / Nominal defocus min: 0.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)