+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | SARS-CoV-2 Spike Hexapro - C68.59 Fab (Class 2 - Fab bound) | |||||||||
 Map data Map data | Sars-CoV-2 Spike Hexapro C59.68 Fab (Class 2 - Fab bound) | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | spike / fusion / membrane / VIRAL PROTEIN | |||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | |||||||||
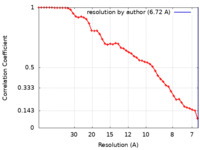
| Method | single particle reconstruction / cryo EM / Resolution: 6.72 Å | |||||||||
 Authors Authors | Croft JT / Lee KK | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: bioRxiv / Year: 2023 Journal: bioRxiv / Year: 2023Title: Identification of broad, potent antibodies to functionally constrained regions of SARS-CoV-2 spike following a breakthrough infection. Authors: Jamie Guenthoer / Michelle Lilly / Tyler N Starr / Bernadeta Dadonaite / Klaus N Lovendahl / Jacob T Croft / Caitlin I Stoddard / Vrasha Chohan / Shilei Ding / Felicitas Ruiz / Mackenzie S ...Authors: Jamie Guenthoer / Michelle Lilly / Tyler N Starr / Bernadeta Dadonaite / Klaus N Lovendahl / Jacob T Croft / Caitlin I Stoddard / Vrasha Chohan / Shilei Ding / Felicitas Ruiz / Mackenzie S Kopp / Andr S Finzi / Jesse D Bloom / Helen Y Chu / Kelly K Lee / Julie Overbaugh Abstract: The antiviral benefit of antibodies can be compromised by viral escape especially for rapidly evolving viruses. Therefore, durable, effective antibodies must be both broad and potent to counter newly ...The antiviral benefit of antibodies can be compromised by viral escape especially for rapidly evolving viruses. Therefore, durable, effective antibodies must be both broad and potent to counter newly emerging, diverse strains. Discovery of such antibodies is critically important for SARS-CoV-2 as the global emergence of new variants of concern (VOC) has compromised the efficacy of therapeutic antibodies and vaccines. We describe a collection of broad and potent neutralizing monoclonal antibodies (mAbs) isolated from an individual who experienced a breakthrough infection with the Delta VOC. Four mAbs potently neutralize the Wuhan-Hu-1 vaccine strain, the Delta VOC, and also retain potency against the Omicron VOCs through BA.4/BA.5 in both pseudovirus-based and authentic virus assays. Three mAbs also retain potency to recently circulating VOCs XBB.1.5 and BQ.1.1 and one also potently neutralizes SARS-CoV-1. The potency of these mAbs was greater against Omicron VOCs than all but one of the mAbs that had been approved for therapeutic applications. The mAbs target distinct epitopes on the spike glycoprotein, three in the receptor binding domain (RBD) and one in an invariant region downstream of the RBD in subdomain 1 (SD1). The escape pathways we defined at single amino acid resolution with deep mutational scanning show they target conserved, functionally constrained regions of the glycoprotein, suggesting escape could incur a fitness cost. Overall, these mAbs are novel in their breadth across VOCs, their epitope specificity, and include a highly potent mAb targeting a rare epitope outside of the RBD in SD1. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_29054.map.gz emd_29054.map.gz | 4.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-29054-v30.xml emd-29054-v30.xml emd-29054.xml emd-29054.xml | 19.6 KB 19.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_29054_fsc.xml emd_29054_fsc.xml | 5.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_29054.png emd_29054.png | 23.6 KB | ||
| Filedesc metadata |  emd-29054.cif.gz emd-29054.cif.gz | 6.4 KB | ||
| Others |  emd_29054_half_map_1.map.gz emd_29054_half_map_1.map.gz emd_29054_half_map_2.map.gz emd_29054_half_map_2.map.gz | 4.9 MB 4.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-29054 http://ftp.pdbj.org/pub/emdb/structures/EMD-29054 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29054 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29054 | HTTPS FTP |
-Validation report
| Summary document |  emd_29054_validation.pdf.gz emd_29054_validation.pdf.gz | 591.4 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_29054_full_validation.pdf.gz emd_29054_full_validation.pdf.gz | 591 KB | Display | |
| Data in XML |  emd_29054_validation.xml.gz emd_29054_validation.xml.gz | 9.8 KB | Display | |
| Data in CIF |  emd_29054_validation.cif.gz emd_29054_validation.cif.gz | 13.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29054 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29054 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29054 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29054 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_29054.map.gz / Format: CCP4 / Size: 6.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_29054.map.gz / Format: CCP4 / Size: 6.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sars-CoV-2 Spike Hexapro C59.68 Fab (Class 2 - Fab bound) | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 3.372 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Sars-CoV-2 Spike Hexapro C59.68 Fab (Class 2 - Fab bound) half map 1
| File | emd_29054_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sars-CoV-2 Spike Hexapro C59.68 Fab (Class 2 - Fab bound) half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Sars-CoV-2 Spike Hexapro C59.68 Fab (Class 2 - Fab bound) half map 1
| File | emd_29054_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sars-CoV-2 Spike Hexapro C59.68 Fab (Class 2 - Fab bound) half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Sars-CoV-2 Hexapro
| Entire | Name: Sars-CoV-2 Hexapro |
|---|---|
| Components |
|
-Supramolecule #1: Sars-CoV-2 Hexapro
| Supramolecule | Name: Sars-CoV-2 Hexapro / type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: all Details: Sars-CoV-2 Hexapro, recombinantly expressed and purified. |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: SARS-CoV-2 Spike Hexapro
| Macromolecule | Name: SARS-CoV-2 Spike Hexapro / type: protein_or_peptide / ID: 1 / Enantiomer: DEXTRO |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MFVFLVLLPL VSSQCVNLTT RTQLPPAYTN SFTRGVYYPD KVFRSSVLHS TQDLFLPFFS NVTWFHAIHV SGTNGTKRFD NPVLPFNDGV YFASTEKSNI IRGWIFGTTL DSKTQSLLIV NNATNVVIKV CEFQFCNDPF LGVYYHKNNK SWMESEFRVY SSANNCTFEY ...String: MFVFLVLLPL VSSQCVNLTT RTQLPPAYTN SFTRGVYYPD KVFRSSVLHS TQDLFLPFFS NVTWFHAIHV SGTNGTKRFD NPVLPFNDGV YFASTEKSNI IRGWIFGTTL DSKTQSLLIV NNATNVVIKV CEFQFCNDPF LGVYYHKNNK SWMESEFRVY SSANNCTFEY VSQPFLMDLE GKQGNFKNLR EFVFKNIDGY FKIYSKHTPI NLVRDLPQGF SALEPLVDLP IGINITRFQT LLALHRSYLT PGDSSSGWTA GAAAYYVGYL QPRTFLLKYN ENGTITDAVD CALDPLSETK CTLKSFTVEK GIYQTSNFRV QPTESIVRFP NITNLCPFGE VFNATRFASV YAWNRKRISN CVADYSVLYN SASFSTFKCY GVSPTKLNDL CFTNVYADSF VIRGDEVRQI APGQTGKIAD YNYKLPDDFT GCVIAWNSNN LDSKVGGNYN YLYRLFRKSN LKPFERDIST EIYQAGSTPC NGVEGFNCYF PLQSYGFQPT NGVGYQPYRV VVLSFELLHA PATVCGPKKS TNLVKNKCVN FNFNGLTGTG VLTESNKKFL PFQQFGRDIA DTTDAVRDPQ TLEILDITPC SFGGVSVITP GTNTSNQVAV LYQDVNCTEV PVAIHADQLT PTWRVYSTGS NVFQTRAGCL IGAEHVNNSY ECDIPIGAGI CASYQTQTNS PGSASSVASQ SIIAYTMSLG AENSVAYSNN SIAIPTNFTI SVTTEILPVS MTKTSVDCTM YICGDSTECS NLLLQYGSFC TQLNRALTGI AVEQDKNTQE VFAQVKQIYK TPPIKDFGGF NFSQILPDPS KPSKRSPIED LLFNKVTLAD AGFIKQYGDC LGDIAARDLI CAQKFNGLTV LPPLLTDEMI AQYTSALLAG TITSGWTFGA GPALQIPFPM QMAYRFNGIG VTQNVLYENQ KLIANQFNSA IGKIQDSLSS TPSALGKLQD VVNQNAQALN TLVKQLSSNF GAISSVLNDI LSRLDPPEAE VQIDRLITGR LQSLQTYVTQ QLIRAAEIRA SANLAATKMS ECVLGQSKRV DFCGKGYHLM SFPQSAPHGV VFLHVTYVPA QEKNFTTAPA ICHDGKAHFP REGVFVSNGT HWFVTQRNFY EPQIITTDNT FVSGNCDVVI GIVNNTVYDP LQPELDSFKE ELDKYFKNHT SPDVDLGDIS GINASVVNIQ KEIDRLNEVA KNLNESLIDL QELGKYEQGS GYIPEAPRDG QAYVRKDGEW VLLSTFLGRS LEVLFQGPGS GGGRGVPHIV MVDAYKRYKG GGSAWSHPQF EKGGGSGGGS GGSAWSHPQF EK |
-Macromolecule #2: C68.59 Fab Heavy Chain
| Macromolecule | Name: C68.59 Fab Heavy Chain / type: protein_or_peptide / ID: 2 / Enantiomer: DEXTRO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: EVQLVESGGG LIQPGGSLRL SCAASGLTFT NYWMLWVRQA PGKGLVWVSH INSDGSSTNN ADSVKGRFT ISRDNAKNTL YLQMNSLRDE DTAVYYCGGT YCTGGSCGIV YWGQGTLVTV S S |
-Macromolecule #3: C68.59 Fab Light Chain
| Macromolecule | Name: C68.59 Fab Light Chain / type: protein_or_peptide / ID: 3 / Enantiomer: DEXTRO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: YELTQPPSVS VSPGQTASIT CSGDKLGDKY VFWYQQKSGQ SPALVIYQDS LRPSGIPDRF SGSNAGNTA TLTISGTPAV DEADYYCQAW DSSTLVFGGG TKLTVL |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2.25 mg/mL |
|---|---|
| Buffer | pH: 7.5 / Details: HEPES Buffered Saline: 50 mM HEPES, 280 mM NaCl |
| Grid | Model: Quantifoil R2/2 / Support film - Material: CARBON / Support film - topology: HOLEY / Support film - Film thickness: 10 |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 295.0 K / Instrument: FEI VITROBOT MARK IV / Details: blot force 0, blot time 4s. |
| Details | Combined with 17 uM C68.59 FAb (this class of particles did not display FAb density |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Number grids imaged: 1 / Number real images: 3043 / Average exposure time: 2.397 sec. / Average electron dose: 50.0 e/Å2 Details: Images collected in movie mode at 0.03 second frames in super-resolution mode |
| Electron beam | Acceleration voltage: 300 kV / Electron source: OTHER |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: OTHER / Nominal defocus max: 1.2 µm / Nominal defocus min: 0.7000000000000001 µm / Nominal magnification: 105000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)