[English] 日本語
 Yorodumi
Yorodumi- EMDB-28890: Cryo-EM structure of alkane 1-monooxygenase AlkB-AlkG complex fro... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of alkane 1-monooxygenase AlkB-AlkG complex from Fontimonas thermophila | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Electron transfer complex / iron-sulfur cluster / histidine-diiron center / hydrophobic alkane binding pocket / OXIDOREDUCTASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationmonooxygenase activity / lipid metabolic process / iron ion binding / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Fontimonas thermophila (bacteria) Fontimonas thermophila (bacteria) | |||||||||
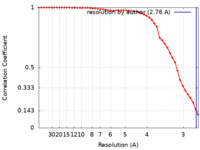
| Method | single particle reconstruction / cryo EM / Resolution: 2.76 Å | |||||||||
 Authors Authors | Chai J / Guo G / McSweeney S / Shanklin J / Liu Q | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2023 Journal: Nat Struct Mol Biol / Year: 2023Title: Structural basis for enzymatic terminal C-H bond functionalization of alkanes. Authors: Jin Chai / Gongrui Guo / Sean M McSweeney / John Shanklin / Qun Liu /  Abstract: Alkane monooxygenase (AlkB) is a widely occurring integral membrane metalloenzyme that catalyzes the initial step in the functionalization of recalcitrant alkanes with high terminal selectivity. AlkB ...Alkane monooxygenase (AlkB) is a widely occurring integral membrane metalloenzyme that catalyzes the initial step in the functionalization of recalcitrant alkanes with high terminal selectivity. AlkB enables diverse microorganisms to use alkanes as their sole carbon and energy source. Here we present the 48.6-kDa cryo-electron microscopy structure of a natural fusion from Fontimonas thermophila between AlkB and its electron donor AlkG at 2.76 Å resolution. The AlkB portion contains six transmembrane helices with an alkane entry tunnel within its transmembrane domain. A dodecane substrate is oriented by hydrophobic tunnel-lining residues to present a terminal C-H bond toward a diiron active site. AlkG, an [Fe-4S] rubredoxin, docks via electrostatic interactions and sequentially transfers electrons to the diiron center. The archetypal structural complex presented reveals the basis for terminal C-H selectivity and functionalization within this broadly distributed evolutionary class of enzymes. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_28890.map.gz emd_28890.map.gz | 4.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-28890-v30.xml emd-28890-v30.xml emd-28890.xml emd-28890.xml | 14 KB 14 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_28890_fsc.xml emd_28890_fsc.xml | 4.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_28890.png emd_28890.png | 122.8 KB | ||
| Filedesc metadata |  emd-28890.cif.gz emd-28890.cif.gz | 5.4 KB | ||
| Others |  emd_28890_half_map_1.map.gz emd_28890_half_map_1.map.gz emd_28890_half_map_2.map.gz emd_28890_half_map_2.map.gz | 7.4 MB 7.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-28890 http://ftp.pdbj.org/pub/emdb/structures/EMD-28890 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28890 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28890 | HTTPS FTP |
-Validation report
| Summary document |  emd_28890_validation.pdf.gz emd_28890_validation.pdf.gz | 818.5 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_28890_full_validation.pdf.gz emd_28890_full_validation.pdf.gz | 818.1 KB | Display | |
| Data in XML |  emd_28890_validation.xml.gz emd_28890_validation.xml.gz | 11 KB | Display | |
| Data in CIF |  emd_28890_validation.cif.gz emd_28890_validation.cif.gz | 13.8 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28890 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28890 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28890 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28890 | HTTPS FTP |
-Related structure data
| Related structure data |  8f6tMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_28890.map.gz / Format: CCP4 / Size: 8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_28890.map.gz / Format: CCP4 / Size: 8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.332 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_28890_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
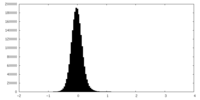
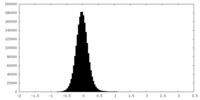
| Density Histograms |
-Half map: #1
| File | emd_28890_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
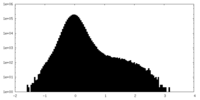
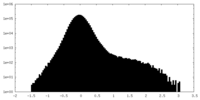
| Density Histograms |
- Sample components
Sample components
-Entire : Electron transfer complex of AlkB and AlkG
| Entire | Name: Electron transfer complex of AlkB and AlkG |
|---|---|
| Components |
|
-Supramolecule #1: Electron transfer complex of AlkB and AlkG
| Supramolecule | Name: Electron transfer complex of AlkB and AlkG / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Fontimonas thermophila (bacteria) Fontimonas thermophila (bacteria) |
-Macromolecule #1: Alkane 1-monooxygenase
| Macromolecule | Name: Alkane 1-monooxygenase / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number: alkane 1-monooxygenase |
|---|---|
| Source (natural) | Organism:  Fontimonas thermophila (bacteria) Fontimonas thermophila (bacteria) |
| Molecular weight | Theoretical: 52.942164 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: SNAMSTPTLD AGTLAWNDGK RYLWLLSPFI PVLGLIGLGL FLYTDIGLFT WSGPLLIYGL IPLLDWLVGE DRNNPPEAAV AQLENDRYY RAIVYAYLPT QYAVTVLGTW VAVTADLAIW EYIGLVLSVG AVNGIGINTA HELGHKRENL DRWLAKLTLA P VAYGHFFV ...String: SNAMSTPTLD AGTLAWNDGK RYLWLLSPFI PVLGLIGLGL FLYTDIGLFT WSGPLLIYGL IPLLDWLVGE DRNNPPEAAV AQLENDRYY RAIVYAYLPT QYAVTVLGTW VAVTADLAIW EYIGLVLSVG AVNGIGINTA HELGHKRENL DRWLAKLTLA P VAYGHFFV EHNRGHHKNV ATPEDPASSK MGESFWAFLP RTVIGSLRSA WAIEKARLQR NKQSVWSLDN ENLQAWAMTI VL FGALTAC LGWPALLFLV LQAAYGASLL EVINYIEHYG LLRQKLPDGR YERCQPRHSW NSNHIVTNLF LYQLQRHSDH HAN PTRRFQ ALRHFDDSPQ LPSGYASMLI PAYVPWLWFR LMDPLVARHY GGDLTKANLY PPKRAALLAR WHRPRPDARR ADTQ PTDAT ATPADAAASP GGRYQCTDCG YIYDEAIGCP REGFPPGTPW SQIPDDWSCP DCAVRDKVDF RKLPAA UniProtKB: Alkane 1-monooxygenase |
-Macromolecule #2: FE (III) ION
| Macromolecule | Name: FE (III) ION / type: ligand / ID: 2 / Number of copies: 3 / Formula: FE |
|---|---|
| Molecular weight | Theoretical: 55.845 Da |
-Macromolecule #3: DODECANE
| Macromolecule | Name: DODECANE / type: ligand / ID: 3 / Number of copies: 1 / Formula: D12 |
|---|---|
| Molecular weight | Theoretical: 170.335 Da |
| Chemical component information |  ChemComp-D12: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.6 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 66.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.7000000000000001 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)