+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of a Zinc-loaded wild-type YiiP-Fab complex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Zinc transporter / cation diffusion facilitator / membrane protein / transport protein | |||||||||
| Function / homology |  Function and homology information Function and homology informationzinc efflux active transmembrane transporter activity / cadmium ion transmembrane transporter activity / ferrous iron transmembrane transporter activity / intracellular zinc ion homeostasis / metal ion binding / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Shewanella oneidensis (bacteria) / Shewanella oneidensis (bacteria) /  Homo sapiens (human) Homo sapiens (human) | |||||||||
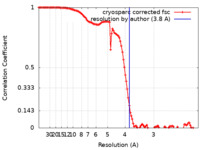
| Method | single particle reconstruction / cryo EM / Resolution: 3.8 Å | |||||||||
 Authors Authors | Lopez-Redondo ML / Hussein AK / Stokes DL | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Elife / Year: 2023 Journal: Elife / Year: 2023Title: Energy coupling and stoichiometry of Zn/H antiport by the prokaryotic cation diffusion facilitator YiiP. Authors: Adel Hussein / Shujie Fan / Maria Lopez-Redondo / Ian Kenney / Xihui Zhang / Oliver Beckstein / David L Stokes /  Abstract: YiiP from Shewanella oneidensis is a prokaryotic Zn/H antiporter that serves as a model for the Cation Diffusion Facilitator (CDF) superfamily, members of which are generally responsible for ...YiiP from Shewanella oneidensis is a prokaryotic Zn/H antiporter that serves as a model for the Cation Diffusion Facilitator (CDF) superfamily, members of which are generally responsible for homeostasis of transition metal ions. Previous studies of YiiP as well as related CDF transporters have established a homodimeric architecture and the presence of three distinct Zn binding sites named A, B, and C. In this study, we use cryo-EM, microscale thermophoresis and molecular dynamics simulations to address the structural and functional roles of individual sites as well as the interplay between Zn binding and protonation. Structural studies indicate that site C in the cytoplasmic domain is primarily responsible for stabilizing the dimer and that site B at the cytoplasmic membrane surface controls the structural transition from an inward facing conformation to an occluded conformation. Binding data show that intramembrane site A, which is directly responsible for transport, has a dramatic pH dependence consistent with coupling to the proton motive force. A comprehensive thermodynamic model encompassing Zn binding and protonation states of individual residues indicates a transport stoichiometry of 1 Zn to 2-3 H depending on the external pH. This stoichiometry would be favorable in a physiological context, allowing the cell to use the proton gradient as well as the membrane potential to drive the export of Zn. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_28881.map.gz emd_28881.map.gz | 129.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-28881-v30.xml emd-28881-v30.xml emd-28881.xml emd-28881.xml | 22.3 KB 22.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_28881_fsc.xml emd_28881_fsc.xml | 11.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_28881.png emd_28881.png | 40.8 KB | ||
| Masks |  emd_28881_msk_1.map emd_28881_msk_1.map | 137.1 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-28881.cif.gz emd-28881.cif.gz | 7 KB | ||
| Others |  emd_28881_half_map_1.map.gz emd_28881_half_map_1.map.gz emd_28881_half_map_2.map.gz emd_28881_half_map_2.map.gz | 127.4 MB 127.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-28881 http://ftp.pdbj.org/pub/emdb/structures/EMD-28881 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28881 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28881 | HTTPS FTP |
-Validation report
| Summary document |  emd_28881_validation.pdf.gz emd_28881_validation.pdf.gz | 749.9 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_28881_full_validation.pdf.gz emd_28881_full_validation.pdf.gz | 749.4 KB | Display | |
| Data in XML |  emd_28881_validation.xml.gz emd_28881_validation.xml.gz | 19.5 KB | Display | |
| Data in CIF |  emd_28881_validation.cif.gz emd_28881_validation.cif.gz | 25.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28881 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28881 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28881 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28881 | HTTPS FTP |
-Related structure data
| Related structure data |  8f6eMC  8f6fC  8f6hC  8f6iC  8f6jC  8f6kC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_28881.map.gz / Format: CCP4 / Size: 137.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_28881.map.gz / Format: CCP4 / Size: 137.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.068 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_28881_msk_1.map emd_28881_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_28881_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_28881_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Wild-type Zinc-loaded YiiP-Fab complex
| Entire | Name: Wild-type Zinc-loaded YiiP-Fab complex |
|---|---|
| Components |
|
-Supramolecule #1: Wild-type Zinc-loaded YiiP-Fab complex
| Supramolecule | Name: Wild-type Zinc-loaded YiiP-Fab complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:  Shewanella oneidensis (bacteria) Shewanella oneidensis (bacteria) |
| Molecular weight | Theoretical: 163.47 KDa |
-Macromolecule #1: Cadmium and zinc efflux pump FieF
| Macromolecule | Name: Cadmium and zinc efflux pump FieF / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Shewanella oneidensis (bacteria) Shewanella oneidensis (bacteria) |
| Molecular weight | Theoretical: 32.485211 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MTQTSQYDFW VKLASRASVA TALTLITIKL LAWLYSGSAS MLASLTDSFA DTLASIINFI AIRYAIVPAD HDHRYGHGKA EPLAALAQS AFIMGSAFLL LFYGGERLLN PSPVENATLG VVVSVVAIVL TLALVLLQKR ALAATNSTVV EADSLHYKSD L FLNAAVLL ...String: MTQTSQYDFW VKLASRASVA TALTLITIKL LAWLYSGSAS MLASLTDSFA DTLASIINFI AIRYAIVPAD HDHRYGHGKA EPLAALAQS AFIMGSAFLL LFYGGERLLN PSPVENATLG VVVSVVAIVL TLALVLLQKR ALAATNSTVV EADSLHYKSD L FLNAAVLL ALVLSQYGWW WADGLFAVLI ACYIGQQAFD LGYRSIQALL DRELDEDTRQ RIKLIAKEDP RVLGLHDLRT RQ AGKTVFI QFHLELDGNL SLNEAHSITD TTGLRVKAAF EDAEVIIHQD PVQVEPTTQ UniProtKB: Cation-efflux pump FieF |
-Macromolecule #2: Fab light chain
| Macromolecule | Name: Fab light chain / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 23.580242 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: SDIQMTQSPS SLSASVGDRV TITCRASQSV SSAVAWYQQK PGKAPKLLIY SASSLYSGVP SRFSGSRSGT DFTLTISSLQ PEDFATYYC QQIWSWPLIT FGQGTKVEIK RTVAAPSVFI FPPSDSQLKS GTASVVCLLN NFYPREAKVQ WKVDNALQSG N SQESVTEQ ...String: SDIQMTQSPS SLSASVGDRV TITCRASQSV SSAVAWYQQK PGKAPKLLIY SASSLYSGVP SRFSGSRSGT DFTLTISSLQ PEDFATYYC QQIWSWPLIT FGQGTKVEIK RTVAAPSVFI FPPSDSQLKS GTASVVCLLN NFYPREAKVQ WKVDNALQSG N SQESVTEQ DSKDSTYSLS STLTLSKADY EKHKVYACEV THQGLSSPVT KSFNRGEC |
-Macromolecule #3: Fab heavy chain
| Macromolecule | Name: Fab heavy chain / type: protein_or_peptide / ID: 3 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 25.406352 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: EISEVQLVES GGGLVQPGGS LRLSCAASGF TIYSSSIHWV RQAPGKGLEW VASIYSSSGS TYYADSVKGR FTISADTSKN TAYLQMNSL RAEDTAVYYC ARQSYSGLSP RRHWSYGAMD YWGQGTLVTV FNQIKGPSVF PLAPSSKSTS GGTAALGCLV K DYFPEPVT ...String: EISEVQLVES GGGLVQPGGS LRLSCAASGF TIYSSSIHWV RQAPGKGLEW VASIYSSSGS TYYADSVKGR FTISADTSKN TAYLQMNSL RAEDTAVYYC ARQSYSGLSP RRHWSYGAMD YWGQGTLVTV FNQIKGPSVF PLAPSSKSTS GGTAALGCLV K DYFPEPVT VSWNSGALTS GVHTFPAVLQ SSGLYSLSSV VTVPSSSLGT QTYICNVNHK PSNTKVDKKV EPKSCDKTHT |
-Macromolecule #4: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 4 / Number of copies: 8 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 4 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| |||||||||||||||
| Grid | Model: C-flat-1.2/1.3 / Material: COPPER / Mesh: 400 / Support film - Material: CARBON / Support film - topology: HOLEY ARRAY / Support film - Film thickness: 7 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 5 sec. / Pretreatment - Atmosphere: AIR / Pretreatment - Pressure: 0.02 kPa / Details: PELCO easiGlow | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Detector mode: SUPER-RESOLUTION / Digitization - Dimensions - Width: 5760 pixel / Digitization - Dimensions - Height: 4092 pixel / Number grids imaged: 2 / Number real images: 7205 / Average exposure time: 2.5 sec. / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 81000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)