[English] 日本語
 Yorodumi
Yorodumi- EMDB-28672: Single-Molecule 3D Image of 16 Helix RNA Origami Satellite by Ind... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Single-Molecule 3D Image of 16 Helix RNA Origami Satellite by Individual Particle Electron Tomography (No. 04) | ||||||||||||||||||
 Map data Map data | 3D Image of 16 Helix RNA Origami Satellite by Individual Particle Electron Tomography (No. 04) | ||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||
 Keywords Keywords | RNA / origami / Individual Particle Electron Tomography / Single-Molecule 3D Image | ||||||||||||||||||
| Biological species | unidentified (others) | ||||||||||||||||||
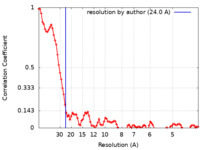
| Method | electron tomography / cryo EM / Resolution: 24.0 Å | ||||||||||||||||||
 Authors Authors | Liu J / Ren G | ||||||||||||||||||
| Funding support |  United States, 5 items United States, 5 items
| ||||||||||||||||||
 Citation Citation |  Journal: Nat Nanotechnol / Year: 2023 Journal: Nat Nanotechnol / Year: 2023Title: Structure, folding and flexibility of co-transcriptional RNA origami. Authors: Ewan K S McRae / Helena Østergaard Rasmussen / Jianfang Liu / Andreas Bøggild / Michael T A Nguyen / Nestor Sampedro Vallina / Thomas Boesen / Jan Skov Pedersen / Gang Ren / Cody Geary / ...Authors: Ewan K S McRae / Helena Østergaard Rasmussen / Jianfang Liu / Andreas Bøggild / Michael T A Nguyen / Nestor Sampedro Vallina / Thomas Boesen / Jan Skov Pedersen / Gang Ren / Cody Geary / Ebbe Sloth Andersen /   Abstract: RNA origami is a method for designing RNA nanostructures that can self-assemble through co-transcriptional folding with applications in nanomedicine and synthetic biology. However, to advance the ...RNA origami is a method for designing RNA nanostructures that can self-assemble through co-transcriptional folding with applications in nanomedicine and synthetic biology. However, to advance the method further, an improved understanding of RNA structural properties and folding principles is required. Here we use cryogenic electron microscopy to study RNA origami sheets and bundles at sub-nanometre resolution revealing structural parameters of kissing-loop and crossover motifs, which are used to improve designs. In RNA bundle designs, we discover a kinetic folding trap that forms during folding and is only released after 10 h. Exploration of the conformational landscape of several RNA designs reveal the flexibility of helices and structural motifs. Finally, sheets and bundles are combined to construct a multidomain satellite shape, which is characterized by individual-particle cryo-electron tomography to reveal the domain flexibility. Together, the study provides a structural basis for future improvements to the design cycle of genetically encoded RNA nanodevices. | ||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_28672.map.gz emd_28672.map.gz | 58.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-28672-v30.xml emd-28672-v30.xml emd-28672.xml emd-28672.xml | 13.6 KB 13.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_28672_fsc.xml emd_28672_fsc.xml | 10.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_28672.png emd_28672.png | 27.6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-28672 http://ftp.pdbj.org/pub/emdb/structures/EMD-28672 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28672 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28672 | HTTPS FTP |
-Validation report
| Summary document |  emd_28672_validation.pdf.gz emd_28672_validation.pdf.gz | 276 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_28672_full_validation.pdf.gz emd_28672_full_validation.pdf.gz | 275.5 KB | Display | |
| Data in XML |  emd_28672_validation.xml.gz emd_28672_validation.xml.gz | 11.3 KB | Display | |
| Data in CIF |  emd_28672_validation.cif.gz emd_28672_validation.cif.gz | 14.8 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28672 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28672 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28672 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28672 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_28672.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_28672.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | 3D Image of 16 Helix RNA Origami Satellite by Individual Particle Electron Tomography (No. 04) | ||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.1 Å | ||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : 16 Helix RNA origami satellite
| Entire | Name: 16 Helix RNA origami satellite |
|---|---|
| Components |
|
-Supramolecule #1: 16 Helix RNA origami satellite
| Supramolecule | Name: 16 Helix RNA origami satellite / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all / Details: In vitro purified RNA. |
|---|---|
| Source (natural) | Organism: unidentified (others) |
| Molecular weight | Theoretical: 590 KDa |
-Macromolecule #1: 16 Helix RNA origami satellite
| Macromolecule | Name: 16 Helix RNA origami satellite / type: rna / ID: 1 |
|---|---|
| Sequence | String: GGAACAAGCA UAACCCGUCC AGGUUCUCAA GACCAAAGAG AACUUGGACC UAUAGUCGGC GUCUACCCGU GUCGCCACGG GUGGACGCCG GCUAUAGGCU UCCCUUUCAU CGCACCUCGC GUUCGCACGC GGGGUGCGCU GACGGUGGGG CUAGCGCCCC AUCGUCAGCC ...String: GGAACAAGCA UAACCCGUCC AGGUUCUCAA GACCAAAGAG AACUUGGACC UAUAGUCGGC GUCUACCCGU GUCGCCACGG GUGGACGCCG GCUAUAGGCU UCCCUUUCAU CGCACCUCGC GUUCGCACGC GGGGUGCGCU GACGGUGGGG CUAGCGCCCC AUCGUCAGCC CUAUCCCGUU ACUUUACAGA GGUCCCGCGC AAUUUGUGGA CCUUGGCAGG UCCGCAAAUU GCGUGGGACG GUGCGGGGUU UCAUGGCUGA AACCUCGCAC CGUAAUAGCG GUGAUUAACG AGCUAGCGCU CGUUGAUCAC CCACUUCGGU AACUCAAGAA CCGGAGUGCG UUUGGAUCUU CAAUCCUAGA GAAGGUCCAA ACGGCUGUUA CCUCUGUGAA GUAACGACGC GUCACAAUGG ACUAGUGAUG CGUCGCACGC UACCCCAAAA CCAGAGGGGU GGCGUGCGGG GUAGGGAUGA AGGGGAAGCG GGUUGUGUUU GUUCCCAAAU ACCUUCUUCG GGCCUCUCUU CCAUGAUACU AUUGAUUAUU UCGCUUCUAA UAUACCCCGU AGUCUCCGGA CUACGCCCGA CGUCCGCGUC GGGGAGGGGC UAACAGACUA AGCUCCUCGG CCCGUAAUAC CGAAACGGGG UAAUACCCAC CCGCAGUUCC CGGGACUGUA AUCGGUAAAC AGUCGGCGCA AAGUCUGAGC GCCCUUACUC UUCGGAGUAA GCCGGCAGUA UACGUACUGC CCGAGGGUGC UCGUAUUUGU GCGCAAAUAC CGCAUAUGGU ACGCCAUAUG GGAGUGGUGA CUUUCGAGUC AUCACUCCCA CGUUUGAAGG AUGAACAAGC GUGCGCCCUU GAAAUCGUCA CAAGGGGAGC GCCCUCGGGG AAUUGCGGCG GUCACAAGAC GAUAGUGACG GCCCAAUCAU CCAGGGCCCC AACUCUACGG AGUUGGCGCU AGUCUUACGA GACUAGGUGG GUGUUACGUA UAUUGGAAGC GAGAUAAUCA AUGGUAUCCU GGUUAAUGGA GGAGAGGGGC GUUUUAUAGU GAAAGUCCAA CACUAUGAAA CCGGGCUCCG GGAUCAACUA GGAAGAUCUC GGAGCGGCGC AUUUUCUAUU CGCCUUCUCU UACAUAUCUA AAUACUCUUU UCUCGCGCUG GUAUCCGUAC CAGCCGGAUG AUACGUCAUC CGCAGCUGAG AAUAGGUGAC UCAGCUGGCG CGGAAAUCCU GUAUCCGCCU UACUUCCGCC UACGCUCUAG GCGUGGAUAA ACAGGAAAUC CACGACCCAA CACCUAAGGG UCGAGCAAUU CCGAUUGCUC GCCCUCGAAU ACGUUCGAGG CUGGCGUGCU GGGUGGUGUC CGCACCACCG GGAUCCUGUA CGCAGGAUCG GCGAGGAGGG UUUCGACCCU UCUCGCCGAG UGAAUAACUG UUGAAUUCGC UCCCCGUCCC AAGUCUUGAG GGACGCAGCA UGCCAGCUAG GGCGUAGCGG CCUUAACAAG ACAAAGGCGA GGUAACAACA GAACCUCCCC UGUCUGCGGA CAGGGCGGUU GCUCUCCGGA GCAACGCGGA GGUAAGGAGA GAAGAGUGUU UAGAUGUGUA AGAGGAGGCG CUUGAGAAAU AGAGAAUGCC AUAGCCGUAU GUAAAUUGGU CAUACAUAUG GCUAUGGGUC UGCAGUUACG ACUGCGGACC GCCCUGAUGU AGGUACGCCU AUAUCAGCCG GGCUGCUCCG UACGCGGAGU AGCCGCCGUC GGGUGGCUAC GGCCAUCCGA CCCCGCAGUC AUGAUUCGUC AUGGCUGCCA GACGUGCGGA UUACGAUCCG UACGUCUGGA AGGAGGUGUU UG |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | electron tomography |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.8 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Chamber temperature: 277 K / Instrument: LEICA EM GP | ||||||||||||
| Details | In vitro transcribed RNA was purified by size exclusion chromatography and spin concentrated in amicon spin columns. | ||||||||||||
| Sectioning | Other: NO SECTIONING |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 6.2 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 2.0 µm / Nominal magnification: 42000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)