[English] 日本語
 Yorodumi
Yorodumi- EMDB-28646: EM map of the human UBR5 HECT-type E3 ubiquitin ligase in a tetra... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
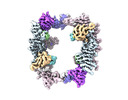
| Title | EM map of the human UBR5 HECT-type E3 ubiquitin ligase in a tetrameric form | |||||||||
 Map data Map data | The final composite map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | HECT / E3 ligase / LIGASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationheterochromatin boundary formation / protein K29-linked ubiquitination / cytoplasm protein quality control by the ubiquitin-proteasome system / protein branched polyubiquitination / nuclear protein quality control by the ubiquitin-proteasome system / HECT-type E3 ubiquitin transferase / cytoplasm protein quality control / protein K11-linked ubiquitination / ubiquitin-ubiquitin ligase activity / DNA repair-dependent chromatin remodeling ...heterochromatin boundary formation / protein K29-linked ubiquitination / cytoplasm protein quality control by the ubiquitin-proteasome system / protein branched polyubiquitination / nuclear protein quality control by the ubiquitin-proteasome system / HECT-type E3 ubiquitin transferase / cytoplasm protein quality control / protein K11-linked ubiquitination / ubiquitin-ubiquitin ligase activity / DNA repair-dependent chromatin remodeling / protein K48-linked ubiquitination / progesterone receptor signaling pathway / negative regulation of smoothened signaling pathway / ubiquitin binding / positive regulation of protein import into nucleus / protein polyubiquitination / ubiquitin protein ligase activity / positive regulation of canonical Wnt signaling pathway / proteasome-mediated ubiquitin-dependent protein catabolic process / DNA repair / DNA damage response / positive regulation of gene expression / chromatin / perinuclear region of cytoplasm / protein-containing complex / RNA binding / zinc ion binding / nucleoplasm / membrane / nucleus / cytosol / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
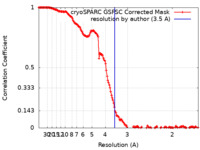
| Method | single particle reconstruction / cryo EM / Resolution: 3.5 Å | |||||||||
 Authors Authors | Wang F / He Q / Lin G / Li H | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Structure / Year: 2023 Journal: Structure / Year: 2023Title: Structure of the human UBR5 E3 ubiquitin ligase. Authors: Feng Wang / Qing He / Wenhu Zhan / Ziqi Yu / Efrat Finkin-Groner / Xiaojing Ma / Gang Lin / Huilin Li /  Abstract: The human UBR5 is a single polypeptide chain homology to E6AP C terminus (HECT)-type E3 ubiquitin ligase essential for embryonic development in mammals. Dysregulated UBR5 functions like an ...The human UBR5 is a single polypeptide chain homology to E6AP C terminus (HECT)-type E3 ubiquitin ligase essential for embryonic development in mammals. Dysregulated UBR5 functions like an oncoprotein to promote cancer growth and metastasis. Here, we report that UBR5 assembles into a dimer and a tetramer. Our cryoelectron microscopy (cryo-EM) structures reveal that two crescent-shaped UBR5 monomers assemble head to tail to form the dimer, and two dimers bind face to face to form the cage-like tetramer with all four catalytic HECT domains facing the central cavity. Importantly, the N-terminal region of one subunit and the HECT of the other form an "intermolecular jaw" in the dimer. We show the jaw-lining residues are important for function, suggesting that the intermolecular jaw functions to recruit ubiquitin-loaded E2 to UBR5. Further work is needed to understand how oligomerization regulates UBR5 ligase activity. This work provides a framework for structure-based anticancer drug development and contributes to a growing appreciation of E3 ligase diversity. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_28646.map.gz emd_28646.map.gz | 277 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-28646-v30.xml emd-28646-v30.xml emd-28646.xml emd-28646.xml | 21.8 KB 21.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_28646_fsc.xml emd_28646_fsc.xml | 14.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_28646.png emd_28646.png | 88.6 KB | ||
| Filedesc metadata |  emd-28646.cif.gz emd-28646.cif.gz | 7.7 KB | ||
| Others |  emd_28646_additional_1.map.gz emd_28646_additional_1.map.gz emd_28646_half_map_1.map.gz emd_28646_half_map_1.map.gz emd_28646_half_map_2.map.gz emd_28646_half_map_2.map.gz | 159.9 MB 302 MB 302 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-28646 http://ftp.pdbj.org/pub/emdb/structures/EMD-28646 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28646 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28646 | HTTPS FTP |
-Validation report
| Summary document |  emd_28646_validation.pdf.gz emd_28646_validation.pdf.gz | 796.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_28646_full_validation.pdf.gz emd_28646_full_validation.pdf.gz | 796.3 KB | Display | |
| Data in XML |  emd_28646_validation.xml.gz emd_28646_validation.xml.gz | 23.6 KB | Display | |
| Data in CIF |  emd_28646_validation.cif.gz emd_28646_validation.cif.gz | 30.6 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28646 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28646 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28646 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28646 | HTTPS FTP |
-Related structure data
| Related structure data |  8ewiMC  8d4xC  8e0qC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_28646.map.gz / Format: CCP4 / Size: 325 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_28646.map.gz / Format: CCP4 / Size: 325 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | The final composite map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.828 Å | ||||||||||||||||||||||||||||||||||||
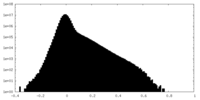
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: Focused on refine map A
| File | emd_28646_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Focused_on_refine_map_A | ||||||||||||
| Projections & Slices |
| ||||||||||||
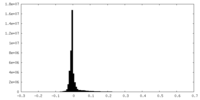
| Density Histograms |
-Half map: Half map a
| File | emd_28646_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half_map_a | ||||||||||||
| Projections & Slices |
| ||||||||||||
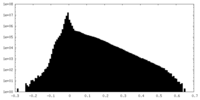
| Density Histograms |
-Half map: Half map B
| File | emd_28646_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half_map_B | ||||||||||||
| Projections & Slices |
| ||||||||||||
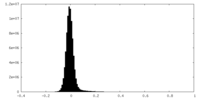
| Density Histograms |
- Sample components
Sample components
-Entire : Homodimer of human E3 ligase UBR5
| Entire | Name: Homodimer of human E3 ligase UBR5 |
|---|---|
| Components |
|
-Supramolecule #1: Homodimer of human E3 ligase UBR5
| Supramolecule | Name: Homodimer of human E3 ligase UBR5 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 / Details: UBR5 expressed in insect cell |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 610 KDa |
-Macromolecule #1: E3 ubiquitin-protein ligase UBR5
| Macromolecule | Name: E3 ubiquitin-protein ligase UBR5 / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO / EC number: HECT-type E3 ubiquitin transferase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 309.72275 KDa |
| Recombinant expression | Organism: Insect cell expression vector pTIE1 (others) |
| Sequence | String: MTSIHFVVHP LPGTEDQLND RLREVSEKLN KYNLNSHPPL NVLEQATIKQ CVVGPNHAAF LLEDGRVCRI GFSVQPDRLE LGKPDNNDG SKLNSNSGAG RTSRPGRTSD SPWFLSGSET LGRLAGNTLG SRWSSGVGGS GGGSSGRSSA GARDSRRQTR V IRTGRDRG ...String: MTSIHFVVHP LPGTEDQLND RLREVSEKLN KYNLNSHPPL NVLEQATIKQ CVVGPNHAAF LLEDGRVCRI GFSVQPDRLE LGKPDNNDG SKLNSNSGAG RTSRPGRTSD SPWFLSGSET LGRLAGNTLG SRWSSGVGGS GGGSSGRSSA GARDSRRQTR V IRTGRDRG SGLLGSQPQP VIPASVIPEE LISQAQVVLQ GKSRSVIIRE LQRTNLDVNL AVNNLLSRDD EDGDDGDDTA SE SYLPGED LMSLLDADIH SAHPSVIIDA DAMFSEDISY FGYPSFRRSS LSRLGSSRVL LLPLERDSEL LRERESVLRL RER RWLDGA SFDNERGSTS KEGEPNLDKK NTPVQSPVSL GEDLQWWPDK DGTKFICIGA LYSELLAVSS KGELYQWKWS ESEP YRNAQ NPSLHHPRAT FLGLTNEKIV LLSANSIRAT VATENNKVAT WVDETLSSVA SKLEHTAQTY SELQGERIVS LHCCA LYTC AQLENSLYWW GVVPFSQRKK MLEKARAKNK KPKSSAGISS MPNITVGTQV CLRNNPLYHA GAVAFSISAG IPKVGV LME SVWNMNDSCR FQLRSPESLK NMEKASKTTE AKPESKQEPV KTEMGPPPSP ASTCSDASSI ASSASMPYKR RRSTPAP KE EEKVNEEQWS LREVVFVEDV KNVPVGKVLK VDGAYVAVKF PGTSSNTNCQ NSSGPDADPS SLLQDCRLLR IDELQVVK T GGTPKVPDCF QRTPKKLCIP EKTEILAVNV DSKGVHAVLK TGNWVRYCIF DLATGKAEQE NNFPTSSIAF LGQNERNVA IFTAGQESPI ILRDGNGTIY PMAKDCMGGI RDPDWLDLPP ISSLGMGVHS LINLPANSTI KKKAAVIIMA VEKQTLMQHI LRCDYEACR QYLMNLEQAV VLEQNLQMLQ TFISHRCDGN RNILHACVSV CFPTSNKETK EEEEAERSER NTFAERLSAV E AIANAISV VSSNGPGNRA GSSSSRSLRL REMMRRSLRA AGLGRHEAGA SSSDHQDPVS PPIAPPSWVP DPPAMDPDGD ID FILAPAV GSLTTAATGT GQGPSTSTIP GPSTEPSVVE SKDRKANAHF ILKLLCDSVV LQPYLRELLS AKDARGMTPF MSA VSGRAY PAAITILETA QKIAKAEISS SEKEEDVFMG MVCPSGTNPD DSPLYVLCCN DTCSFTWTGA EHINQDIFEC RTCG LLESL CCCTECARVC HKGHDCKLKR TSPTAYCDCW EKCKCKTLIA GQKSARLDLL YRLLTATNLV TLPNSRGEHL LLFLV QTVA RQTVEHCQYR PPRIREDRNR KTASPEDSDM PDHDLEPPRF AQLALERVLQ DWNALKSMIM FGSQENKDPL SASSRI GHL LPEEQVYLNQ QSGTIRLDCF THCLIVKCTA DILLLDTLLG TLVKELQNKY TPGRREEAIA VTMRFLRSVA RVFVILS VE MASSKKKNNF IPQPIGKCKR VFQALLPYAV EELCNVAESL IVPVRMGIAR PTAPFTLAST SIDAMQGSEE LFSVEPLP P RPSSDQSSSS SQSQSSYIIR NPQQRRISQS QPVRGRDEEQ DDIVSADVEE VEVVEGVAGE EDHHDEQEEH GEENAEAEG QHDEHDEDGS DMELDLLAAA ETESDSESNH SNQDNASGRR SVVTAATAGS EAGASSVPAF FSEDDSQSND SSDSDSSSSQ SDDIEQETF MLDEPLERTT NSSHANGAAQ APRSMQWAVR NTQHQRAAST APSSTSTPAA SSAGLIYIDP SNLRRSGTIS T SAAAAAAA LEASNASSYL TSASSLARAY SIVIRQISDL MGLIPKYNHL VYSQIPAAVK LTYQDAVNLQ NYVEEKLIPT WN WMVSIMD STEAQLRYGS ALASAGDPGH PNHPLHASQN SARRERMTAR EEASLRTLEG RRRATLLSAR QGMMSARGDF LNY ALSLMR SHNDEHSDVL PVLDVCSLKH VAYVFQALIY WIKAMNQQTT LDTPQLERKR TRELLELGID NEDSEHENDD DTNQ SATLN DKDDDSLPAE TGQNHPFFRR SDSMTFLGCI PPNPFEVPLA EAIPLADQPH LLQPNARKED LFGRPSQGLY SSSAS SGKC LMEVTVDRNC LEVLPTKMSY AANLKNVMNM QNRQKKEGEE QPVLPEETES SKPGPSAHDL AAQLKSSLLA EIGLTE SEG PPLTSFRPQC SFMGMVISHD MLLGRWRLSL ELFGRVFMED VGAEPGSILT ELGGFEVKES KFRREMEKLR NQQSRDL SL EVDRDRDLLI QQTMRQLNNH FGRRCATTPM AVHRVKVTFK DEPGEGSGVA RSFYTAIAQA FLSNEKLPNL ECIQNANK G THTSLMQRLR NRGERDRERE REREMRRSSG LRAGSRRDRD RDFRRQLSID TRPFRPASEG NPSDDPEPLP AHRQALGER LYPRVQAMQP AFASKITGML LELSPAQLLL LLASEDSLRA RVDEAMELII AHGRENGADS ILDLGLVDSS EKVQQENRKR HGSSRSVVD MDLDDTDDGD DNAPLFYQPG KRGFYTPRPG KNTEARLNCF RNIGRILGLC LLQNELCPIT LNRHVIKVLL G RKVNWHDF AFFDPVMYES LRQLILASQS SDADAVFSAM DLAFAIDLCK EEGGGQVELI PNGVNIPVTP QNVYEYVRKY AE HRMLVVA EQPLHAMRKG LLDVLPKNSL EDLTAEDFRL LVNGCGEVNV QMLISFTSFN DESGENAEKL LQFKRWFWSI VEK MSMTER QDLVYFWTSS PSLPASEEGF QPMPSITIRP PDDQHLPTAN TCISRLYVPL YSSKQILKQK LLLAIKTKNF GFV UniProtKB: E3 ubiquitin-protein ligase UBR5 |
-Macromolecule #2: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 2 / Number of copies: 12 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.7 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| ||||||||||||
| Grid | Model: Quantifoil R2/1 / Support film - Material: GOLD / Support film - topology: CONTINUOUS | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 279 K / Instrument: FEI VITROBOT MARK IV / Details: blot 2S, blot forth 2. | ||||||||||||
| Details | freshly purified UBR5 |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Temperature | Min: 193.0 K / Max: 193.0 K |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 65.0 e/Å2 Details: Images were collected in movie-mode at 75 frames per second |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 105000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: RIGID BODY FIT / Overall B value: 119 / Target criteria: Correlation coefficient |
|---|---|
| Output model |  PDB-8ewi: |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)