[English] 日本語
 Yorodumi
Yorodumi- EMDB-28605: Acheta domesticus segmented densovirus high buoyancy fraction 1 (... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
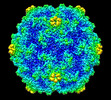
| Title | Acheta domesticus segmented densovirus high buoyancy fraction 1 (HB1) empty capsid structure | |||||||||
 Map data Map data | Acheta domesticus segmented densovirus high buoyancy fraction 1 (HB1) empty capsid structure | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | parvovirus / densovirus / virus capsid / virion / insect virus / capsid / invertebrate / virus / VIRUS LIKE PARTICLE | |||||||||
| Function / homology | Capsid protein Function and homology information Function and homology information | |||||||||
| Biological species |  Solenopsis invicta-associated densovirus / Solenopsis invicta-associated densovirus /  unclassified Densovirinae (virus) unclassified Densovirinae (virus) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.0 Å | |||||||||
 Authors Authors | Penzes JJ / McKenna R / Tijssen P | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Bipartite genome and structural organization of the parvovirus Acheta domesticus segmented densovirus. Authors: Judit J Pénzes / Hanh T Pham / Paul Chipman / Emmanuel W Smith / Robert McKenna / Peter Tijssen /   Abstract: Parvoviruses (family Parvoviridae) are currently defined by a linear monopartite ssDNA genome, T = 1 icosahedral capsids, and distinct structural (VP) and non-structural (NS) protein expression ...Parvoviruses (family Parvoviridae) are currently defined by a linear monopartite ssDNA genome, T = 1 icosahedral capsids, and distinct structural (VP) and non-structural (NS) protein expression cassettes within their genome. We report the discovery of a parvovirus with a bipartite genome, Acheta domesticus segmented densovirus (AdSDV), isolated from house crickets (Acheta domesticus), in which it is pathogenic. We found that the AdSDV harbors its NS and VP cassettes on two separate genome segments. Its vp segment acquired a phospholipase A2-encoding gene, vpORF3, via inter-subfamily recombination, coding for a non-structural protein. We showed that the AdSDV evolved a highly complex transcription profile in response to its multipartite replication strategy compared to its monopartite ancestors. Our structural and molecular examinations revealed that the AdSDV packages one genome segment per particle. The cryo-EM structures of two empty- and one full-capsid population (3.3, 3.1 and 2.3 Å resolution) reveal a genome packaging mechanism, which involves an elongated C-terminal tail of the VP, "pinning" the ssDNA genome to the capsid interior at the twofold symmetry axis. This mechanism fundamentally differs from the capsid-DNA interactions previously seen in parvoviruses. This study provides new insights on the mechanism behind ssDNA genome segmentation and on the plasticity of parvovirus biology. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_28605.map.gz emd_28605.map.gz | 443.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-28605-v30.xml emd-28605-v30.xml emd-28605.xml emd-28605.xml | 12.4 KB 12.4 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_28605.png emd_28605.png | 211 KB | ||
| Filedesc metadata |  emd-28605.cif.gz emd-28605.cif.gz | 5.9 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-28605 http://ftp.pdbj.org/pub/emdb/structures/EMD-28605 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28605 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28605 | HTTPS FTP |
-Validation report
| Summary document |  emd_28605_validation.pdf.gz emd_28605_validation.pdf.gz | 561.5 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_28605_full_validation.pdf.gz emd_28605_full_validation.pdf.gz | 561.1 KB | Display | |
| Data in XML |  emd_28605_validation.xml.gz emd_28605_validation.xml.gz | 8.2 KB | Display | |
| Data in CIF |  emd_28605_validation.cif.gz emd_28605_validation.cif.gz | 9.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28605 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28605 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28605 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28605 | HTTPS FTP |
-Related structure data
| Related structure data |  8eu6MC  8er8C  8erkC  8eu7C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_28605.map.gz / Format: CCP4 / Size: 476.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_28605.map.gz / Format: CCP4 / Size: 476.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Acheta domesticus segmented densovirus high buoyancy fraction 1 (HB1) empty capsid structure | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.059 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : unclassified Densovirinae
| Entire | Name:  unclassified Densovirinae (virus) unclassified Densovirinae (virus) |
|---|---|
| Components |
|
-Supramolecule #1: unclassified Densovirinae
| Supramolecule | Name: unclassified Densovirinae / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all / NCBI-ID: 185880 / Sci species name: unclassified Densovirinae / Virus type: VIRION / Virus isolate: STRAIN / Virus enveloped: No / Virus empty: Yes |
|---|---|
| Host (natural) | Organism:  Acheta domesticus (house cricket) Acheta domesticus (house cricket) |
-Macromolecule #1: Capsid protein
| Macromolecule | Name: Capsid protein / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Solenopsis invicta-associated densovirus Solenopsis invicta-associated densovirus |
| Molecular weight | Theoretical: 36.291906 KDa |
| Recombinant expression | Organism:  Acheta domesticus (house cricket) Acheta domesticus (house cricket) |
| Sequence | String: EGYGKHITSM HVRNIFNQGN QVIRNIVKQQ RYELLDFTGT EAGTTNLPKI IPYQCIWWRG LQNAANVNQT INNMIALNTI SYGVRFLKA KLCIEVYAVT RKRLIQTGAT SYYTDDFEQG QNLFIGWADR KAESIPITTP ADLDETKLTV ANTTLFDANN D NITKEEVP ...String: EGYGKHITSM HVRNIFNQGN QVIRNIVKQQ RYELLDFTGT EAGTTNLPKI IPYQCIWWRG LQNAANVNQT INNMIALNTI SYGVRFLKA KLCIEVYAVT RKRLIQTGAT SYYTDDFEQG QNLFIGWADR KAESIPITTP ADLDETKLTV ANTTLFDANN D NITKEEVP TREKWCHTWD LDVLNHNYLW EPNNLDSQWT LIPGAQAVQP TATPIGPTYQ EIVIATKAIG ANESALVTTI QD RRSYPRL MLSQPQIKDE TDTMKFKYQI RISTELEMEH HIKPDIANPW LTRQTLPLPA LSGDGTTRYV PCVPYETHVS Q UniProtKB: Capsid protein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL |
|---|---|
| Buffer | pH: 7.4 / Component - Concentration: 1.0 x / Component - Name: Phosphate buffered saline |
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 45 sec. |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Instrument: FEI VITROBOT MARK IV |
| Details | Deceased common house crickets, previously showing signs of paralysis, inability to jump, and inability to control movement |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Phase plate: VOLTA PHASE PLATE |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Number grids imaged: 1 / Number real images: 970 / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source: OTHER |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: OTHER / Nominal defocus max: 4.0 µm / Nominal defocus min: 0.81 µm |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) X (Row.)
X (Row.) Y (Col.)
Y (Col.)




















