+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM map of octopus sensory receptor CRT1 | |||||||||
 Map data Map data | Cryo-EM structure of octopus sensory receptor | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | pentameric ligand gated ion channel / octopus sensory receptor / cys-loop receptor / STRUCTURAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationextracellular ligand-gated monoatomic ion channel activity / transmembrane signaling receptor activity / postsynapse / membrane Similarity search - Function | |||||||||
| Biological species |  Octopus bimaculoides (California two-spot octopus) Octopus bimaculoides (California two-spot octopus) | |||||||||
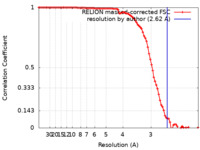
| Method | single particle reconstruction / cryo EM / Resolution: 2.62 Å | |||||||||
 Authors Authors | Kang G / Kim JJ / Allard CAH / Valencia-Montoya WA / Bellono NW / Hibbs RE | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2023 Journal: Nature / Year: 2023Title: Sensory specializations drive octopus and squid behaviour. Authors: Guipeun Kang / Corey A H Allard / Wendy A Valencia-Montoya / Lena van Giesen / Jeong Joo Kim / Peter B Kilian / Xiaochen Bai / Nicholas W Bellono / Ryan E Hibbs /  Abstract: The evolution of new traits enables expansion into new ecological and behavioural niches. Nonetheless, demonstrated connections between divergence in protein structure, function and lineage-specific ...The evolution of new traits enables expansion into new ecological and behavioural niches. Nonetheless, demonstrated connections between divergence in protein structure, function and lineage-specific behaviours remain rare. Here we show that both octopus and squid use cephalopod-specific chemotactile receptors (CRs) to sense their respective marine environments, but structural adaptations in these receptors support the sensation of specific molecules suited to distinct physiological roles. We find that squid express ancient CRs that more closely resemble related nicotinic acetylcholine receptors, whereas octopuses exhibit a more recent expansion in CRs consistent with their elaborated 'taste by touch' sensory system. Using a combination of genetic profiling, physiology and behavioural analyses, we identify the founding member of squid CRs that detects soluble bitter molecules that are relevant in ambush predation. We present the cryo-electron microscopy structure of a squid CR and compare this with octopus CRs and nicotinic receptors. These analyses demonstrate an evolutionary transition from an ancestral aromatic 'cage' that coordinates soluble neurotransmitters or tastants to a more recent octopus CR hydrophobic binding pocket that traps insoluble molecules to mediate contact-dependent chemosensation. Thus, our study provides a foundation for understanding how adaptation of protein structure drives the diversification of organismal traits and behaviour. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_28163.map.gz emd_28163.map.gz | 40.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-28163-v30.xml emd-28163-v30.xml emd-28163.xml emd-28163.xml | 15 KB 15 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_28163_fsc.xml emd_28163_fsc.xml | 9.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_28163.png emd_28163.png | 91.1 KB | ||
| Filedesc metadata |  emd-28163.cif.gz emd-28163.cif.gz | 5.6 KB | ||
| Others |  emd_28163_half_map_1.map.gz emd_28163_half_map_1.map.gz emd_28163_half_map_2.map.gz emd_28163_half_map_2.map.gz | 52.1 MB 52.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-28163 http://ftp.pdbj.org/pub/emdb/structures/EMD-28163 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28163 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28163 | HTTPS FTP |
-Validation report
| Summary document |  emd_28163_validation.pdf.gz emd_28163_validation.pdf.gz | 723.6 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_28163_full_validation.pdf.gz emd_28163_full_validation.pdf.gz | 723.1 KB | Display | |
| Data in XML |  emd_28163_validation.xml.gz emd_28163_validation.xml.gz | 16.3 KB | Display | |
| Data in CIF |  emd_28163_validation.cif.gz emd_28163_validation.cif.gz | 21.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28163 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28163 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28163 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28163 | HTTPS FTP |
-Related structure data
| Related structure data |  8eisMC  8eizC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_28163.map.gz / Format: CCP4 / Size: 67 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_28163.map.gz / Format: CCP4 / Size: 67 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM structure of octopus sensory receptor | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.056 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Half Map 1
| File | emd_28163_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half Map 2
| File | emd_28163_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Octopus sensory receptor CRT1 in complex with diosgenin
| Entire | Name: Octopus sensory receptor CRT1 in complex with diosgenin |
|---|---|
| Components |
|
-Supramolecule #1: Octopus sensory receptor CRT1 in complex with diosgenin
| Supramolecule | Name: Octopus sensory receptor CRT1 in complex with diosgenin type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Octopus bimaculoides (California two-spot octopus) Octopus bimaculoides (California two-spot octopus) |
| Molecular weight | Theoretical: 200 kDa/nm |
-Macromolecule #1: Octopus sensory receptor
| Macromolecule | Name: Octopus sensory receptor / type: protein_or_peptide / ID: 1 / Number of copies: 5 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Octopus bimaculoides (California two-spot octopus) Octopus bimaculoides (California two-spot octopus) |
| Molecular weight | Theoretical: 43.768008 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: TPTYGDERLL REKLLTNYSK SIRPVINLTK VVDVTALLYL QTLYDLDFVN NFIMARYYLG LIWIDEKLTW NPLDYNNITS IYLPKDKIW TPPIKMCNSM DKSEENDGVG ELMLTYTGWI NMWSFRLLHT YCQINAYTYP FDEHTCEIYL CVALHTINHT R IKELIYED ...String: TPTYGDERLL REKLLTNYSK SIRPVINLTK VVDVTALLYL QTLYDLDFVN NFIMARYYLG LIWIDEKLTW NPLDYNNITS IYLPKDKIW TPPIKMCNSM DKSEENDGVG ELMLTYTGWI NMWSFRLLHT YCQINAYTYP FDEHTCEIYL CVALHTINHT R IKELIYED SKFTQNYKWD INVSGKVNGT DELFSYAFAP MYLRRKLTVG IIAMLIPTVM MTILTIFVFL LPPESGEKVS LA TTIFLSN VLYLVQIDKT TPTNTKYPSL LMLYLMLLSM LSGIATLGSV VISKLYVIQS PSLRKSNPSD QNMNKSHTNK VAD ISTISK VQSDLPIREK PNEKRIYCIS DYIRLDDIFL KLSIATSVII SMIFTCLLFI PLE UniProtKB: Neurotransmitter-gated ion-channel ligand-binding domain-containing protein |
-Macromolecule #2: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 2 / Number of copies: 20 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Macromolecule #3: 2-[2-[(1~{S},2~{S},4~{S},5'~{R},6~{R},7~{S},8~{R},9~{S},12~{S},13...
| Macromolecule | Name: 2-[2-[(1~{S},2~{S},4~{S},5'~{R},6~{R},7~{S},8~{R},9~{S},12~{S},13~{R},16~{S})-5',7,9,13-tetramethylspiro[5-oxapentacyclo[10.8.0.0^{2,9}.0^{4,8}.0^{13,18}]icos-18-ene-6,2'-oxane]-16-yl]oxyethyl]propane-1,3-diol type: ligand / ID: 3 / Number of copies: 5 / Formula: DU0 |
|---|---|
| Molecular weight | Theoretical: 516.752 Da |
| Chemical component information |  ChemComp-DU0: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)