[English] 日本語
 Yorodumi
Yorodumi- EMDB-28160: The complex of phosphorylated human delta F508 cystic fibrosis tr... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | The complex of phosphorylated human delta F508 cystic fibrosis transmembrane conductance regulator (CFTR) with elexacaftor (VX-445), lumacaftor (VX-809) and ATP/Mg | |||||||||
 Map data Map data | Delta 508 cystic fibrosis transmembrane conductance regulator (d508CFTR) in complex with lumacaftor, elexacaftor and ATP/Mg | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | ABC transporter / ion channel / folding correction / membrane protein / ATP-Binding Protein | |||||||||
| Function / homology |  Function and homology information Function and homology informationpositive regulation of voltage-gated chloride channel activity / positive regulation of cyclic nucleotide-gated ion channel activity / Sec61 translocon complex binding / channel-conductance-controlling ATPase / intracellularly ATP-gated chloride channel activity / positive regulation of enamel mineralization / transepithelial water transport / RHO GTPases regulate CFTR trafficking / intracellular pH elevation / amelogenesis ...positive regulation of voltage-gated chloride channel activity / positive regulation of cyclic nucleotide-gated ion channel activity / Sec61 translocon complex binding / channel-conductance-controlling ATPase / intracellularly ATP-gated chloride channel activity / positive regulation of enamel mineralization / transepithelial water transport / RHO GTPases regulate CFTR trafficking / intracellular pH elevation / amelogenesis / ATPase-coupled inorganic anion transmembrane transporter activity / chloride channel inhibitor activity / Golgi-associated vesicle membrane / multicellular organismal-level water homeostasis / membrane hyperpolarization / cholesterol transport / bicarbonate transport / bicarbonate transmembrane transporter activity / vesicle docking involved in exocytosis / chloride channel regulator activity / chloride transmembrane transporter activity / sperm capacitation / chloride channel activity / RHOQ GTPase cycle / cholesterol biosynthetic process / positive regulation of exocytosis / positive regulation of insulin secretion involved in cellular response to glucose stimulus / chloride channel complex / ATPase-coupled transmembrane transporter activity / ABC-type transporter activity / cellular response to cAMP / cellular response to forskolin / chloride transmembrane transport / isomerase activity / response to endoplasmic reticulum stress / establishment of localization in cell / PDZ domain binding / Defective CFTR causes cystic fibrosis / Late endosomal microautophagy / clathrin-coated endocytic vesicle membrane / ABC-family proteins mediated transport / recycling endosome / transmembrane transport / Aggrephagy / Chaperone Mediated Autophagy / recycling endosome membrane / Cargo recognition for clathrin-mediated endocytosis / Clathrin-mediated endocytosis / early endosome membrane / protein-folding chaperone binding / early endosome / endosome membrane / Ub-specific processing proteases / apical plasma membrane / lysosomal membrane / endoplasmic reticulum membrane / enzyme binding / cell surface / ATP hydrolysis activity / protein-containing complex / ATP binding / membrane / nucleus / plasma membrane / cytoplasm / cytosol Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
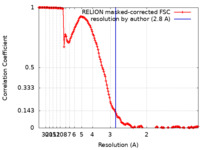
| Method | single particle reconstruction / cryo EM / Resolution: 2.8 Å | |||||||||
 Authors Authors | Fiedorczuk K / Chen J | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Science / Year: 2022 Journal: Science / Year: 2022Title: Molecular structures reveal synergistic rescue of Δ508 CFTR by Trikafta modulators. Authors: Karol Fiedorczuk / Jue Chen /  Abstract: The predominant mutation causing cystic fibrosis, a deletion of phenylalanine 508 (Δ508) in the cystic fibrosis transmembrane conductance regulator (CFTR), leads to severe defects in CFTR biogenesis ...The predominant mutation causing cystic fibrosis, a deletion of phenylalanine 508 (Δ508) in the cystic fibrosis transmembrane conductance regulator (CFTR), leads to severe defects in CFTR biogenesis and function. The advanced therapy Trikafta combines the folding corrector tezacaftor (VX-661), the channel potentiator ivacaftor (VX-770), and the dual-function modulator elexacaftor (VX-445). However, it is unclear how elexacaftor exerts its effects, in part because the structure of Δ508 CFTR is unknown. Here, we present cryo-electron microscopy structures of Δ508 CFTR in the absence and presence of CFTR modulators. When used alone, elexacaftor partially rectified interdomain assembly defects in Δ508 CFTR, but when combined with a type I corrector, did so fully. These data illustrate how the different modulators in Trikafta synergistically rescue Δ508 CFTR structure and function. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_28160.map.gz emd_28160.map.gz | 304.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-28160-v30.xml emd-28160-v30.xml emd-28160.xml emd-28160.xml | 19 KB 19 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_28160_fsc.xml emd_28160_fsc.xml | 15.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_28160.png emd_28160.png | 105 KB | ||
| Masks |  emd_28160_msk_1.map emd_28160_msk_1.map | 325 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-28160.cif.gz emd-28160.cif.gz | 6.8 KB | ||
| Others |  emd_28160_half_map_1.map.gz emd_28160_half_map_1.map.gz emd_28160_half_map_2.map.gz emd_28160_half_map_2.map.gz | 260 MB 260.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-28160 http://ftp.pdbj.org/pub/emdb/structures/EMD-28160 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28160 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28160 | HTTPS FTP |
-Validation report
| Summary document |  emd_28160_validation.pdf.gz emd_28160_validation.pdf.gz | 1.1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_28160_full_validation.pdf.gz emd_28160_full_validation.pdf.gz | 1.1 MB | Display | |
| Data in XML |  emd_28160_validation.xml.gz emd_28160_validation.xml.gz | 23.1 KB | Display | |
| Data in CIF |  emd_28160_validation.cif.gz emd_28160_validation.cif.gz | 30.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28160 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28160 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28160 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28160 | HTTPS FTP |
-Related structure data
| Related structure data |  8eioMC  8eigC  8eiqC  8ej1C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_28160.map.gz / Format: CCP4 / Size: 325 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_28160.map.gz / Format: CCP4 / Size: 325 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Delta 508 cystic fibrosis transmembrane conductance regulator (d508CFTR) in complex with lumacaftor, elexacaftor and ATP/Mg | ||||||||||||||||||||
| Voxel size | X=Y=Z: 0.676 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_28160_msk_1.map emd_28160_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map 1
| File | emd_28160_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map 2
| File | emd_28160_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : The complex of phosphorylated human delta F508 cystic fibrosis tr...
| Entire | Name: The complex of phosphorylated human delta F508 cystic fibrosis transmembrane conductance regulator (CFTR) with elexacaftor (VX-445), lumacaftor (VX-809) and ATP/Mg |
|---|---|
| Components |
|
-Supramolecule #1: The complex of phosphorylated human delta F508 cystic fibrosis tr...
| Supramolecule | Name: The complex of phosphorylated human delta F508 cystic fibrosis transmembrane conductance regulator (CFTR) with elexacaftor (VX-445), lumacaftor (VX-809) and ATP/Mg type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 168 KDa |
-Macromolecule #1: Cystic fibrosis transmembrane conductance regulator
| Macromolecule | Name: Cystic fibrosis transmembrane conductance regulator / type: protein_or_peptide / ID: 1 Details: Expressed construct is human CFTR with F508 deletion and E1371Q substitution Number of copies: 1 / Enantiomer: LEVO / EC number: channel-conductance-controlling ATPase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 168.187297 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MQRSPLEKAS VVSKLFFSWT RPILRKGYRQ RLELSDIYQI PSVDSADNLS EKLEREWDRE LASKKNPKLI NALRRCFFWR FMFYGIFLY LGEVTKAVQP LLLGRIIASY DPDNKEERSI AIYLGIGLCL LFIVRTLLLH PAIFGLHHIG MQMRIAMFSL I YKKTLKLS ...String: MQRSPLEKAS VVSKLFFSWT RPILRKGYRQ RLELSDIYQI PSVDSADNLS EKLEREWDRE LASKKNPKLI NALRRCFFWR FMFYGIFLY LGEVTKAVQP LLLGRIIASY DPDNKEERSI AIYLGIGLCL LFIVRTLLLH PAIFGLHHIG MQMRIAMFSL I YKKTLKLS SRVLDKISIG QLVSLLSNNL NKFDEGLALA HFVWIAPLQV ALLMGLIWEL LQASAFCGLG FLIVLALFQA GL GRMMMKY RDQRAGKISE RLVITSEMIE NIQSVKAYCW EEAMEKMIEN LRQTELKLTR KAAYVRYFNS SAFFFSGFFV VFL SVLPYA LIKGIILRKI FTTISFCIVL RMAVTRQFPW AVQTWYDSLG AINKIQDFLQ KQEYKTLEYN LTTTEVVMEN VTAF WEEGF GELFEKAKQN NNNRKTSNGD DSLFFSNFSL LGTPVLKDIN FKIERGQLLA VAGSTGAGKT SLLMVIMGEL EPSEG KIKH SGRISFCSQF SWIMPGTIKE NIIGVSYDEY RYRSVIKACQ LEEDISKFAE KDNIVLGEGG ITLSGGQRAR ISLARA VYK DADLYLLDSP FGYLDVLTEK EIFESCVCKL MANKTRILVT SKMEHLKKAD KILILHEGSS YFYGTFSELQ NLQPDFS SK LMGCDSFDQF SAERRNSILT ETLHRFSLEG DAPVSWTETK KQSFKQTGEF GEKRKNSILN PINSIRKFSI VQKTPLQM N GIEEDSDEPL ERRLSLVPDS EQGEAILPRI SVISTGPTLQ ARRRQSVLNL MTHSVNQGQN IHRKTTASTR KVSLAPQAN LTELDIYSRR LSQETGLEIS EEINEEDLKE CFFDDMESIP AVTTWNTYLR YITVHKSLIF VLIWCLVIFL AEVAASLVVL WLLGNTPLQ DKGNSTHSRN NSYAVIITST SSYYVFYIYV GVADTLLAMG FFRGLPLVHT LITVSKILHH KMLHSVLQAP M STLNTLKA GGILNRFSKD IAILDDLLPL TIFDFIQLLL IVIGAIAVVA VLQPYIFVAT VPVIVAFIML RAYFLQTSQQ LK QLESEGR SPIFTHLVTS LKGLWTLRAF GRQPYFETLF HKALNLHTAN WFLYLSTLRW FQMRIEMIFV IFFIAVTFIS ILT TGEGEG RVGIILTLAM NIMSTLQWAV NSSIDVDSLM RSVSRVFKFI DMPTEGKPTK STKPYKNGQL SKVMIIENSH VKKD DIWPS GGQMTVKDLT AKYTEGGNAI LENISFSISP GQRVGLLGRT GSGKSTLLSA FLRLLNTEGE IQIDGVSWDS ITLQQ WRKA FGVIPQKVFI FSGTFRKNLD PYEQWSDQEI WKVADEVGLR SVIEQFPGKL DFVLVDGGCV LSHGHKQLMC LARSVL SKA KILLLDQPSA HLDPVTYQII RRTLKQAFAD CTVILCEHRI EAMLECQQFL VIEENKVRQY DSIQKLLNER SLFRQAI SP SDRVKLFPHR NSSKCKSKPQ IAALKEETEE EVQDTRL UniProtKB: Cystic fibrosis transmembrane conductance regulator |
-Macromolecule #2: Cystic fibrosis transmembrane conductance regulator
| Macromolecule | Name: Cystic fibrosis transmembrane conductance regulator / type: protein_or_peptide / ID: 2 Details: R-domain of del508CFTR (UNP:P13569). Expressed construct is human CFTR with F508 deletion and E1371Q substitution. Number of copies: 1 / Enantiomer: LEVO / EC number: channel-conductance-controlling ATPase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 1.464797 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) |
-Macromolecule #3: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 3 / Number of copies: 2 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Macromolecule #4: ADENOSINE-5'-TRIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-TRIPHOSPHATE / type: ligand / ID: 4 / Number of copies: 2 / Formula: ATP |
|---|---|
| Molecular weight | Theoretical: 507.181 Da |
| Chemical component information |  ChemComp-ATP: |
-Macromolecule #5: CHOLESTEROL
| Macromolecule | Name: CHOLESTEROL / type: ligand / ID: 5 / Number of copies: 1 / Formula: CLR |
|---|---|
| Molecular weight | Theoretical: 386.654 Da |
| Chemical component information |  ChemComp-CLR: |
-Macromolecule #6: DODECANE
| Macromolecule | Name: DODECANE / type: ligand / ID: 6 / Number of copies: 1 / Formula: D12 |
|---|---|
| Molecular weight | Theoretical: 170.335 Da |
| Chemical component information |  ChemComp-D12: |
-Macromolecule #7: Lumacaftor
| Macromolecule | Name: Lumacaftor / type: ligand / ID: 7 / Number of copies: 1 / Formula: VX8 |
|---|---|
| Molecular weight | Theoretical: 452.407 Da |
| Chemical component information |  ChemComp-VX8: |
-Macromolecule #8: (6P)-N-(1,3-dimethyl-1H-pyrazole-4-sulfonyl)-6-[3-(3,3,3-trifluor...
| Macromolecule | Name: (6P)-N-(1,3-dimethyl-1H-pyrazole-4-sulfonyl)-6-[3-(3,3,3-trifluoro-2,2-dimethylpropoxy)-1H-pyrazol-1-yl]-2-[(4S)-2,2,4-trimethylpyrrolidin-1-yl]pyridine-3-carboxamide type: ligand / ID: 8 / Number of copies: 1 / Formula: WJX |
|---|---|
| Molecular weight | Theoretical: 597.653 Da |
| Chemical component information | 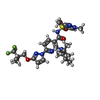 ChemComp-WJX: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 5 mg/mL |
|---|---|
| Buffer | pH: 7.4 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 65.6 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.8 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller









 Z
Z Y
Y X
X