+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM density of BIRC6/casp-3 (clusters) | |||||||||
 Map data Map data | main map / consensus | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Ubiquitin / E3 ligase / Apoptosis / Autophagy / IAP / LIGASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationcaspase-3 / Stimulation of the cell death response by PAK-2p34 / phospholipase A2 activator activity / anterior neural tube closure / intrinsic apoptotic signaling pathway in response to osmotic stress / leukocyte apoptotic process / positive regulation of pyroptotic inflammatory response / glial cell apoptotic process / NADE modulates death signalling / labyrinthine layer development ...caspase-3 / Stimulation of the cell death response by PAK-2p34 / phospholipase A2 activator activity / anterior neural tube closure / intrinsic apoptotic signaling pathway in response to osmotic stress / leukocyte apoptotic process / positive regulation of pyroptotic inflammatory response / glial cell apoptotic process / NADE modulates death signalling / labyrinthine layer development / luteolysis / response to cobalt ion / (E3-independent) E2 ubiquitin-conjugating enzyme / cellular response to staurosporine / death-inducing signaling complex / ALK mutants bind TKIs / cyclin-dependent protein serine/threonine kinase inhibitor activity / Apoptosis induced DNA fragmentation / Apoptotic cleavage of cell adhesion proteins / Caspase activation via Dependence Receptors in the absence of ligand / SMAC, XIAP-regulated apoptotic response / Activation of caspases through apoptosome-mediated cleavage / death receptor binding / Flemming body / Signaling by Hippo / SMAC (DIABLO) binds to IAPs / SMAC(DIABLO)-mediated dissociation of IAP:caspase complexes / axonal fasciculation / regulation of synaptic vesicle cycle / fibroblast apoptotic process / epithelial cell apoptotic process / Other interleukin signaling / platelet formation / negative regulation of cytokine production / positive regulation of amyloid-beta formation / execution phase of apoptosis / negative regulation of B cell proliferation / Apoptotic cleavage of cellular proteins / pyroptotic inflammatory response / negative regulation of activated T cell proliferation / T cell homeostasis / microtubule organizing center / ubiquitin conjugating enzyme activity / neurotrophin TRK receptor signaling pathway / B cell homeostasis / negative regulation of cell cycle / response to tumor necrosis factor / cysteine-type endopeptidase inhibitor activity / cell fate commitment / response to X-ray / response to amino acid / Pyroptosis / regulation of macroautophagy / Caspase-mediated cleavage of cytoskeletal proteins / response to glucose / response to UV / response to glucocorticoid / keratinocyte differentiation / striated muscle cell differentiation / Degradation of the extracellular matrix / intrinsic apoptotic signaling pathway / protein maturation / enzyme activator activity / erythrocyte differentiation / hippocampus development / regulation of cytokinesis / negative regulation of extrinsic apoptotic signaling pathway / response to nicotine / apoptotic signaling pathway / sensory perception of sound / trans-Golgi network / protein catabolic process / protein processing / regulation of protein stability / neuron differentiation / response to hydrogen peroxide / response to wounding / spindle pole / ubiquitin-protein transferase activity / Signaling by ALK fusions and activated point mutants / positive regulation of neuron apoptotic process / response to estradiol / peptidase activity / regulation of cell population proliferation / heart development / protease binding / midbody / neuron apoptotic process / response to lipopolysaccharide / aspartic-type endopeptidase activity / learning or memory / response to hypoxia / cell population proliferation / endosome / postsynaptic density / protein ubiquitination / protein phosphorylation / response to xenobiotic stimulus / cell division / cysteine-type endopeptidase activity Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
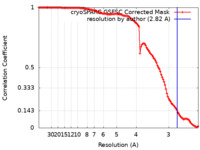
| Method | single particle reconstruction / cryo EM / Resolution: 2.82 Å | |||||||||
 Authors Authors | Hunkeler M / Fischer ES | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Science / Year: 2023 Journal: Science / Year: 2023Title: Structures of BIRC6-client complexes provide a mechanism of SMAC-mediated release of caspases. Authors: Moritz Hunkeler / Cyrus Y Jin / Eric S Fischer /  Abstract: Tight regulation of apoptosis is essential for metazoan development and prevents diseases such as cancer and neurodegeneration. Caspase activation is central to apoptosis, and inhibitor of apoptosis ...Tight regulation of apoptosis is essential for metazoan development and prevents diseases such as cancer and neurodegeneration. Caspase activation is central to apoptosis, and inhibitor of apoptosis proteins (IAPs) are the principal actors that restrain caspase activity and are therefore attractive therapeutic targets. IAPs, in turn, are regulated by mitochondria-derived proapoptotic factors such as SMAC and HTRA2. Through a series of cryo-electron microscopy structures of full-length human baculoviral IAP repeat-containing protein 6 (BIRC6) bound to SMAC, caspases, and HTRA2, we provide a molecular understanding for BIRC6-mediated caspase inhibition and its release by SMAC. The architecture of BIRC6, together with near-irreversible binding of SMAC, elucidates how the IAP inhibitor SMAC can effectively control a processive ubiquitin ligase to respond to apoptotic stimuli. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_27839.map.gz emd_27839.map.gz | 157.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-27839-v30.xml emd-27839-v30.xml emd-27839.xml emd-27839.xml | 39.1 KB 39.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_27839_fsc.xml emd_27839_fsc.xml | 11.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_27839.png emd_27839.png | 109.3 KB | ||
| Masks |  emd_27839_msk_1.map emd_27839_msk_1.map emd_27839_msk_2.map emd_27839_msk_2.map | 166.4 MB 52.7 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-27839.cif.gz emd-27839.cif.gz | 9.4 KB | ||
| Others |  emd_27839_additional_1.map.gz emd_27839_additional_1.map.gz emd_27839_additional_2.map.gz emd_27839_additional_2.map.gz emd_27839_additional_3.map.gz emd_27839_additional_3.map.gz emd_27839_additional_4.map.gz emd_27839_additional_4.map.gz emd_27839_additional_5.map.gz emd_27839_additional_5.map.gz emd_27839_additional_6.map.gz emd_27839_additional_6.map.gz emd_27839_half_map_1.map.gz emd_27839_half_map_1.map.gz emd_27839_half_map_2.map.gz emd_27839_half_map_2.map.gz | 157.1 MB 157.2 MB 157.2 MB 156.8 MB 157.1 MB 157.2 MB 154.2 MB 154.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-27839 http://ftp.pdbj.org/pub/emdb/structures/EMD-27839 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27839 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27839 | HTTPS FTP |
-Validation report
| Summary document |  emd_27839_validation.pdf.gz emd_27839_validation.pdf.gz | 1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_27839_full_validation.pdf.gz emd_27839_full_validation.pdf.gz | 1 MB | Display | |
| Data in XML |  emd_27839_validation.xml.gz emd_27839_validation.xml.gz | 20.1 KB | Display | |
| Data in CIF |  emd_27839_validation.cif.gz emd_27839_validation.cif.gz | 26.1 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27839 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27839 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27839 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27839 | HTTPS FTP |
-Related structure data
| Related structure data |  8e2dC  8e2eC  8e2fC  8e2gC  8e2hC  8e2iC  8e2jC  8e2kC C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_27839.map.gz / Format: CCP4 / Size: 166.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_27839.map.gz / Format: CCP4 / Size: 166.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | main map / consensus | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.2069 Å | ||||||||||||||||||||||||||||||||||||
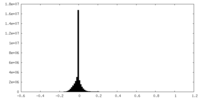
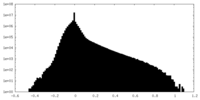
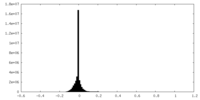
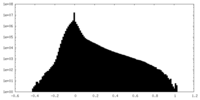
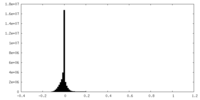
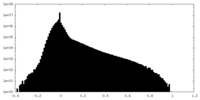
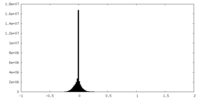
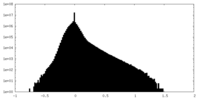
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
+Mask #1
+Mask #2
+Additional map: cluster 1
+Additional map: cluster 3
+Additional map: cluster 2
+Additional map: cluster 4
+Additional map: cluster 5
+Additional map: cluster 6
+Half map: half-map 2
+Half map: half-map 1
- Sample components
Sample components
-Entire : BIRC6/casp-3
| Entire | Name: BIRC6/casp-3 |
|---|---|
| Components |
|
-Supramolecule #1: BIRC6/casp-3
| Supramolecule | Name: BIRC6/casp-3 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 1.132 MDa |
-Macromolecule #1: Baculoviral IAP repeat-containing protein 6
| Macromolecule | Name: Baculoviral IAP repeat-containing protein 6 / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO / EC number: Ligases |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MGDWSHPQFE KSGGGSGGLE VLFQGPSRTM VTGGGAAPPG TVTEPLPSVI VLSAGRKMAA AAAAASGPGC SSAAGAGAAG VSEWLVLRDG CMHCDADGLH SLSYHPALNA ILAVTSRGTI KVIDGTSGAT LQASALSAKP GGQVKCQYIS AVDKVIFVDD YAVGCRKDLN ...String: MGDWSHPQFE KSGGGSGGLE VLFQGPSRTM VTGGGAAPPG TVTEPLPSVI VLSAGRKMAA AAAAASGPGC SSAAGAGAAG VSEWLVLRDG CMHCDADGLH SLSYHPALNA ILAVTSRGTI KVIDGTSGAT LQASALSAKP GGQVKCQYIS AVDKVIFVDD YAVGCRKDLN GILLLDTALQ TPVSKQDDVV QLELPVTEAQ QLLSACLEKV DISSTEGYDL FITQLKDGLK NTSHETAANH KVAKWATVTF HLPHHVLKSI ASAIVNELKK INQNVAALPV ASSVMDRLSY LLPSARPELG VGPGRSVDRS LMYSEANRRE TFTSWPHVGY RWAQPDPMAQ AGFYHQPASS GDDRAMCFTC SVCLVCWEPT DEPWSEHERH SPNCPFVKGE HTQNVPLSVT LATSPAQFPC TDGTDRISCF GSGSCPHFLA AATKRGKICI WDVSKLMKVH LKFEINAYDP AIVQQLILSG DPSSGVDSRR PTLAWLEDSS SCSDIPKLEG DSDDLLEDSD SEEHSRSDSV TGHTSQKEAM EVSLDITALS ILQQPEKLQW EIVANVLEDT VKDLEELGAN PCLTNSKSEK TKEKHQEQHN IPFPCLLAGG LLTYKSPATS PISSNSHRSL DGLSRTQGES ISEQGSTDNE SCTNSELNSP LVRRTLPVLL LYSIKESDEK AGKIFSQMNN IMSKSLHDDG FTVPQIIEME LDSQEQLLLQ DPPVTYIQQF ADAAANLTSP DSEKWNSVFP KPGTLVQCLR LPKFAEEENL CIDSITPCAD GIHLLVGLRT CPVESLSAIN QVEALNNLNK LNSALCNRRK GELESNLAVV NGANISVIQH ESPADVQTPL IIQPEQRNVS GGYLVLYKMN YATRIVTLEE EPIKIQHIKD PQDTITSLIL LPPDILDNRE DDCEEPIEDM QLTSKNGFER EKTSDISTLG HLVITTQGGY VKILDLSNFE ILAKVEPPKK EGTEEQDTFV SVIYCSGTDR LCACTKGGEL HFLQIGGTCD DIDEADILVD GSLSKGIEPS SEGSKPLSNP SSPGISGVDL LVDQPFTLEI LTSLVELTRF ETLTPRFSAT VPPCWVEVQQ EQQQRRHPQH LHQQHHGDAA QHTRTWKLQT DSNSWDEHVF ELVLPKACMV GHVDFKFVLN SNITNIPQIQ VTLLKNKAPG LGKVNALNIE VEQNGKPSLV DLNEEMQHMD VEESQCLRLC PFLEDHKEDI LCGPVWLASG LDLSGHAGML TLTSPKLVKG MAGGKYRSFL IHVKAVNERG TEEICNGGMR PVVRLPSLKH QSNKGYSLAS LLAKVAAGKE KSSNVKNENT SGTRKSENLR GCDLLQEVSV TIRRFKKTSI SKERVQRCAM LQFSEFHEKL VNTLCRKTDD GQITEHAQSL VLDTLCWLAG VHSNGPGSSK EGNENLLSKT RKFLSDIVRV CFFEAGRSIA HKCARFLALC ISNGKCDPCQ PAFGPVLLKA LLDNMSFLPA ATTGGSVYWY FVLLNYVKDE DLAGCSTACA SLLTAVSRQL QDRLTPMEAL LQTRYGLYSS PFDPVLFDLE MSGSSCKNVY NSSIGVQSDE IDLSDVLSGN GKVSSCTAAE GSFTSLTGLL EVEPLHFTCV STSDGTRIER DDAMSSFGVT PAVGGLSSGT VGEASTALSS AAQVALQSLS HAMASAEQQL QVLQEKQQQL LKLQQQKAKL EAKLHQTTAA AAAAASAVGP VHNSVPSNPV AAPGFFIHPS DVIPPTPKTT PLFMTPPLTP PNEAVSVVIN AELAQLFPGS VIDPPAVNLA AHNKNSNKSR MNPLGSGLAL AISHASHFLQ PPPHQSIIIE RMHSGARRFV TLDFGRPILL TDVLIPTCGD LASLSIDIWT LGEEVDGRRL VVATDISTHS LILHDLIPPP VCRFMKITVI GRYGSTNARA KIPLGFYYGH TYILPWESEL KLMHDPLKGE GESANQPEID QHLAMMVALQ EDIQCRYNLA CHRLETLLQS IDLPPLNSAN NAQYFLRKPD KAVEEDSRVF SAYQDCIQLQ LQLNLAHNAV QRLKVALGAS RKMLSETSNP EDLIQTSSTE QLRTIIRYLL DTLLSLLHAS NGHSVPAVLQ STFHAQACEE LFKHLCISGT PKIRLHTGLL LVQLCGGERW WGQFLSNVLQ ELYNSEQLLI FPQDRVFMLL SCIGQRSLSN SGVLESLLNL LDNLLSPLQP QLPMHRRTEG VLDIPMISWV VMLVSRLLDY VATVEDEAAA AKKPLNGNQW SFINNNLHTQ SLNRSSKGSS SLDRLYSRKI RKQLVHHKQQ LNLLKAKQKA LVEQMEKEKI QSNKGSSYKL LVEQAKLKQA TSKHFKDLIR LRRTAEWSRS NLDTEVTTAK ESPEIEPLPF TLAHERCISV VQKLVLFLLS MDFTCHADLL LFVCKVLARI ANATRPTIHL CEIVNEPQLE RLLLLLVGTD FNRGDISWGG AWAQYSLTCM LQDILAGELL APVAAEAMEE GTVGDDVGAT AGDSDDSLQQ SSVQLLETID EPLTHDITGA PPLSSLEKDK EIDLELLQDL MEVDIDPLDI DLEKDPLAAK VFKPISSTWY DYWGADYGTY NYNPYIGGLG IPVAKPPANT EKNGSQTVSV SVSQALDARL EVGLEQQAEL MLKMMSTLEA DSILQALTNT SPTLSQSPTG TDDSLLGGLQ AANQTSQLII QLSSVPMLNV CFNKLFSMLQ VHHVQLESLL QLWLTLSLNS SSTGNKENGA DIFLYNANRI PVISLNQASI TSFLTVLAWY PNTLLRTWCL VLHSLTLMTN MQLNSGSSSA IGTQESTAHL LVSDPNLIHV LVKFLSGTSP HGTNQHSPQV GPTATQAMQE FLTRLQVHLS STCPQIFSEF LLKLIHILST ERGAFQTGQG PLDAQVKLLE FTLEQNFEVV SVSTISAVIE SVTFLVHHYI TCSDKVMSRS GSDSSVGARA CFGGLFANLI RPGDAKAVCG EMTRDQLMFD LLKLVNILVQ LPLSGNREYS ARVSVTTNTT DSVSDEEKVS GGKDGNGSST SVQGSPAYVA DLVLANQQIM SQILSALGLC NSSAMAMIIG ASGLHLTKHE NFHGGLDAIS VGDGLFTILT TLSKKASTVH MMLQPILTYM ACGYMGRQGS LATCQLSEPL LWFILRVLDT SDALKAFHDM GGVQLICNNM VTSTRAIVNT ARSMVSTIMK FLDSGPNKAV DSTLKTRILA SEPDNAEGIH NFAPLGTITS SSPTAQPAEV LLQATPPHRR ARSAAWSYIF LPEEAWCDLT IHLPAAVLLK EIHIQPHLAS LATCPSSVSV EVSADGVNML PLSTPVVTSG LTYIKIQLVK AEVASAVCLR LHRPRDASTL GLSQIKLLGL TAFGTTSSAT VNNPFLPSED QVSKTSIGWL RLLHHCLTHI SDLEGMMASA AAPTANLLQT CAALLMSPYC GMHSPNIEVV LVKIGLQSTR IGLKLIDILL RNCAASGSDP TDLNSPLLFG RLNGLSSDST IDILYQLGTT QDPGTKDRIQ ALLKWVSDSA RVAAMKRSGR MNYMCPNSST VEYGLLMPSP SHLHCVAAIL WHSYELLVEY DLPALLDQEL FELLFNWSMS LPCNMVLKKA VDSLLCSMCH VHPNYFSLLM GWMGITPPPV QCHHRLSMTD DSKKQDLSSS LTDDSKNAQA PLALTESHLA TLASSSQSPE AIKQLLDSGL PSLLVRSLAS FCFSHISSSE SIAQSIDISQ DKLRRHHVPQ QCNKMPITAD LVAPILRFLT EVGNSHIMKD WLGGSEVNPL WTALLFLLCH SGSTSGSHNL GAQQTSARSA SLSSAATTGL TTQQRTAIEN ATVAFFLQCI SCHPNNQKLM AQVLCELFQT SPQRGNLPTS GNISGFIRRL FLQLMLEDEK VTMFLQSPCP LYKGRINATS HVIQHPMYGA GHKFRTLHLP VSTTLSDVLD RVSDTPSITA KLISEQKDDK EKKNHEEKEK VKAENGFQDN YSVVVASGLK SQSKRAVSAT PPRPPSRRGR TIPDKIGSTS GAEAANKIIT VPVFHLFHKL LAGQPLPAEM TLAQLLTLLY DRKLPQGYRS IDLTVKLGSR VITDPSLSKT DSYKRLHPEK DHGDLLASCP EDEALTPGDE CMDGILDESL LETCPIQSPL QVFAGMGGLA LIAERLPMLY PEVIQQVSAP VVTSTTQEKP KDSDQFEWVT IEQSGELVYE APETVAAEPP PIKSAVQTMS PIPAHSLAAF GLFLRLPGYA EVLLKERKHA QCLLRLVLGV TDDGEGSHIL QSPSANVLPT LPFHVLRSLF STTPLTTDDG VLLRRMALEI GALHLILVCL SALSHHSPRV PNSSVNQTEP QVSSSHNPTS TEEQQLYWAK GTGFGTGSTA SGWDVEQALT KQRLEEEHVT CLLQVLASYI NPVSSAVNGE AQSSHETRGQ NSNALPSVLL ELLSQSCLIP AMSSYLRNDS VLDMARHVPL YRALLELLRA IASCAAMVPL LLPLSTENGE EEEEQSECQT SVGTLLAKMK TCVDTYTNRL RSKRENVKTG VKPDASDQEP EGLTLLVPDI QKTAEIVYAA TTSLRQANQE KKLGEYSKKA AMKPKPLSVL KSLEEKYVAV MKKLQFDTFE MVSEDEDGKL GFKVNYHYMS QVKNANDANS AARARRLAQE AVTLSTSLPL SSSSSVFVRC DEERLDIMKV LITGPADTPY ANGCFEFDVY FPQDYPSSPP LVNLETTGGH SVRFNPNLYN DGKVCLSILN TWHGRPEEKW NPQTSSFLQV LVSVQSLILV AEPYFNEPGY ERSRGTPSGT QSSREYDGNI RQATVKWAML EQIRNPSPCF KEVIHKHFYL KRVEIMAQCE EWIADIQQYS SDKRVGRTMS HHAAALKRHT AQLREELLKL PCPEGLDPDT DDAPEVCRAT TGAEETLMHD QVKPSSSKEL PSDFQL UniProtKB: Dual E2 ubiquitin-conjugating enzyme/E3 ubiquitin-protein ligase BIRC6 |
-Macromolecule #2: Caspase-3
| Macromolecule | Name: Caspase-3 / type: protein_or_peptide / ID: 2 / Enantiomer: LEVO / EC number: caspase-3 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  |
| Sequence | String: MENTENSVDS KSIKNLEPKI IHGSESMDSG ISLDNSYKMD YPEMGLCIII NNKNFHKSTG MTSRSGTDVD AANLRETFRN LKYEVRNKND LTREEIVELM RDVSKEDHSK RSSFVCVLLS HGEEGIIFGT NGPVDLKKIT NFFRGDRCRS LTGKPKLFII QACRGTELDC ...String: MENTENSVDS KSIKNLEPKI IHGSESMDSG ISLDNSYKMD YPEMGLCIII NNKNFHKSTG MTSRSGTDVD AANLRETFRN LKYEVRNKND LTREEIVELM RDVSKEDHSK RSSFVCVLLS HGEEGIIFGT NGPVDLKKIT NFFRGDRCRS LTGKPKLFII QACRGTELDC GIETDSGVDD DMACHKIPVE ADFLYAYSTA PGYYSWRNSK DGSWFIQSLC AMLKQYADKL EFMHILTRVN RKVATEFESF SFDATFHAKK QIPCIVSMLT KELYFYHLEH HHHHH UniProtKB: Caspase-3 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
| ||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Support film - Film thickness: 12 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. / Pretreatment - Atmosphere: AIR / Pretreatment - Pressure: 0.039 kPa | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Chamber temperature: 283.15 K / Instrument: LEICA EM GP | ||||||||||||
| Details | added CHAPSO to 0.4 mM |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Details | Data collection in counting mode, using multi-shot scheme (9 holes per stage position, 2 movies per hole) |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 5760 pixel / Digitization - Dimensions - Height: 4092 pixel / Digitization - Frames/image: 1-50 / Number grids imaged: 1 / Number real images: 10422 / Average exposure time: 2.5 sec. / Average electron dose: 51.401 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 105000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: OTHER |
|---|
 Movie
Movie Controller
Controller






























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)