[English] 日本語
 Yorodumi
Yorodumi- EMDB-27819: 3D Reconstruction of PPG Matrix-Landed 20S Proteasome Core Partic... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | 3D Reconstruction of PPG Matrix-Landed 20S Proteasome Core Particle Protein Complexes | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Matrix-Landed / Mass Spectrometry / CELL CYCLE | |||||||||
| Biological species |   Thermoplasma acidophilum (acidophilic) Thermoplasma acidophilum (acidophilic) | |||||||||
| Method | single particle reconstruction / negative staining / Resolution: 15.0 Å | |||||||||
 Authors Authors | Salome AZ / Westphall MS | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Anal Chem / Year: 2022 Journal: Anal Chem / Year: 2022Title: Matrix-Landing Mass Spectrometry for Electron Microscopy Imaging of Native Protein Complexes. Authors: Austin Z Salome / Kenneth W Lee / Timothy Grant / Michael S Westphall / Joshua J Coon /  Abstract: Recently, we described the use of a chemical matrix for landing and preserving the cations of protein-protein complexes within a mass spectrometer (MS) instrument. By use of a glycerol-landing ...Recently, we described the use of a chemical matrix for landing and preserving the cations of protein-protein complexes within a mass spectrometer (MS) instrument. By use of a glycerol-landing matrix, we used negative stain transmission electron microscopy (TEM) to obtain a three-dimensional (3D) reconstruction of landed GroEL complexes. Here, we investigate the utilities of other chemical matrices for their abilities to land, preserve, and allow for direct imaging of these cationic particles using TEM. We report here that poly(propylene) glycol (PPG) offers superior performance over glycerol for matrix landing. We demonstrated the utility of the PPG matrix landing using three protein-protein complexes─GroEL, the 20S proteasome core particle, and β-galactosidase─and obtained a 3D reconstruction of each complex from matrix-landed particles. These structures have no detectable differences from the structures obtained using conventional preparation methods, suggesting the structures are well preserved at least to the resolution limit of the reconstructions (∼20 Å). We conclude that matrix landing offers a direct approach to couple native MS with TEM for protein structure determination. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_27819.map.gz emd_27819.map.gz | 7.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-27819-v30.xml emd-27819-v30.xml emd-27819.xml emd-27819.xml | 12.1 KB 12.1 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_27819.png emd_27819.png | 30.6 KB | ||
| Filedesc metadata |  emd-27819.cif.gz emd-27819.cif.gz | 3.7 KB | ||
| Others |  emd_27819_half_map_1.map.gz emd_27819_half_map_1.map.gz emd_27819_half_map_2.map.gz emd_27819_half_map_2.map.gz | 486.3 KB 486.1 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-27819 http://ftp.pdbj.org/pub/emdb/structures/EMD-27819 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27819 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27819 | HTTPS FTP |
-Validation report
| Summary document |  emd_27819_validation.pdf.gz emd_27819_validation.pdf.gz | 593.1 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_27819_full_validation.pdf.gz emd_27819_full_validation.pdf.gz | 592.7 KB | Display | |
| Data in XML |  emd_27819_validation.xml.gz emd_27819_validation.xml.gz | 9 KB | Display | |
| Data in CIF |  emd_27819_validation.cif.gz emd_27819_validation.cif.gz | 10.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27819 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27819 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27819 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27819 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_27819.map.gz / Format: CCP4 / Size: 8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_27819.map.gz / Format: CCP4 / Size: 8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 3.4 Å | ||||||||||||||||||||||||||||||||||||
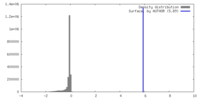
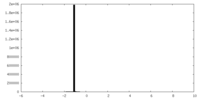
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_27819_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
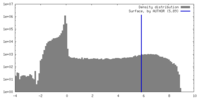
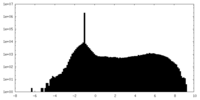
| Density Histograms |
-Half map: #2
| File | emd_27819_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
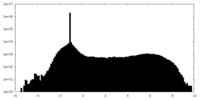
| Density Histograms |
- Sample components
Sample components
-Entire : 20S Proteasome Core Particle
| Entire | Name: 20S Proteasome Core Particle |
|---|---|
| Components |
|
-Supramolecule #1: 20S Proteasome Core Particle
| Supramolecule | Name: 20S Proteasome Core Particle / type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:   Thermoplasma acidophilum (acidophilic) Thermoplasma acidophilum (acidophilic) |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | cell |
- Sample preparation
Sample preparation
| Buffer | pH: 7 |
|---|---|
| Staining | Type: NEGATIVE / Material: Uranyl Acetate |
| Grid | Model: Homemade / Material: COPPER / Mesh: 400 |
| Details | Protein complex solution was ionized and converted to gas-phase via electrospray ionization. Proteasome gas-phase ions were collected for analysis. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI 12 |
|---|---|
| Image recording | Film or detector model: OTHER / Average electron dose: 100.0 e/Å2 / Details: NanoSprint15 MK-II 15 Mpix camera (AMT Imaging) |
| Electron beam | Acceleration voltage: 120 kV / Electron source: LAB6 |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.5 µm |
- Image processing
Image processing
| Startup model | Type of model: OTHER |
|---|---|
| Final reconstruction | Applied symmetry - Point group: D7 (2x7 fold dihedral) / Resolution.type: BY AUTHOR / Resolution: 15.0 Å / Resolution method: OTHER / Software - Name: cisTEM / Number images used: 15695 |
| Initial angle assignment | Type: NOT APPLICABLE |
| Final angle assignment | Type: NOT APPLICABLE |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)