[English] 日本語
 Yorodumi
Yorodumi- EMDB-27717: Cryogenic electron microscopy 3D map of human full-length monomer... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryogenic electron microscopy 3D map of human full-length monomeric alpha-catenin | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | F-actin / cell-cell junction / CELL ADHESION | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
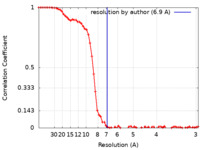
| Method | single particle reconstruction / cryo EM / Resolution: 6.9 Å | |||||||||
 Authors Authors | Rangarajan ES / Izard T | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Commun Biol / Year: 2023 Journal: Commun Biol / Year: 2023Title: Distinct inter-domain interactions of dimeric versus monomeric α-catenin link cell junctions to filaments. Authors: Erumbi S Rangarajan / Emmanuel W Smith / Tina Izard /  Abstract: Attachment between cells is crucial for almost all aspects of the life of cells. These inter-cell adhesions are mediated by the binding of transmembrane cadherin receptors of one cell to cadherins of ...Attachment between cells is crucial for almost all aspects of the life of cells. These inter-cell adhesions are mediated by the binding of transmembrane cadherin receptors of one cell to cadherins of a neighboring cell. Inside the cell, cadherin binds β-catenin, which interacts with α-catenin. The transitioning of cells between migration and adhesion is modulated by α-catenin, which links cell junctions and the plasma membrane to the actin cytoskeleton. At cell junctions, a single β-catenin/α-catenin heterodimer slips along filamentous actin in the direction of cytoskeletal tension which unfolds clustered heterodimers to form catch bonds with F-actin. Outside cell junctions, α-catenin dimerizes and links the plasma membrane to F-actin. Under cytoskeletal tension, α-catenin unfolds and forms an asymmetric catch bond with F-actin. To understand the mechanism of this important α-catenin function, we determined the 2.7 Å cryogenic electron microscopy (cryoEM) structures of filamentous actin alone and bound to human dimeric α-catenin. Our structures provide mechanistic insights into the role of the α-catenin interdomain interactions in directing α-catenin function and suggest a bivalent mechanism. Further, our cryoEM structure of human monomeric α-catenin provides mechanistic insights into α-catenin autoinhibition. Collectively, our structures capture the initial α-catenin interaction with F-actin before the sensing of force, which is a crucial event in cell adhesion and human disease. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_27717.map.gz emd_27717.map.gz | 20.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-27717-v30.xml emd-27717-v30.xml emd-27717.xml emd-27717.xml | 16.5 KB 16.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_27717_fsc.xml emd_27717_fsc.xml | 8.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_27717.png emd_27717.png | 85.5 KB | ||
| Others |  emd_27717_half_map_1.map.gz emd_27717_half_map_1.map.gz emd_27717_half_map_2.map.gz emd_27717_half_map_2.map.gz | 20.7 MB 20.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-27717 http://ftp.pdbj.org/pub/emdb/structures/EMD-27717 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27717 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27717 | HTTPS FTP |
-Validation report
| Summary document |  emd_27717_validation.pdf.gz emd_27717_validation.pdf.gz | 681 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_27717_full_validation.pdf.gz emd_27717_full_validation.pdf.gz | 680.6 KB | Display | |
| Data in XML |  emd_27717_validation.xml.gz emd_27717_validation.xml.gz | 11.7 KB | Display | |
| Data in CIF |  emd_27717_validation.cif.gz emd_27717_validation.cif.gz | 16 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27717 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27717 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27717 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27717 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_27717.map.gz / Format: CCP4 / Size: 22.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_27717.map.gz / Format: CCP4 / Size: 22.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.48 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_27717_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_27717_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Monomeric alpha-catenin (residues 1-906)
| Entire | Name: Monomeric alpha-catenin (residues 1-906) |
|---|---|
| Components |
|
-Supramolecule #1: Monomeric alpha-catenin (residues 1-906)
| Supramolecule | Name: Monomeric alpha-catenin (residues 1-906) / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 100.3 KDa |
-Macromolecule #1: alpha-catenin
| Macromolecule | Name: alpha-catenin / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  |
| Sequence | String: GPLGSMTAVH AGNINFKWDP KSLEIRTLAV ERLLEPLVTQ VTTLVNTNSK GPSNKKRGRS KKAHVLAASV EQATENFLEK GDKIAKESQF LKEELVAAVE DVRKQGDLMK AAAGEFADDP CSSVKRGNMV RAARALLSAV TRLLILADMA DVYKLLVQLK VVEDGILKLR ...String: GPLGSMTAVH AGNINFKWDP KSLEIRTLAV ERLLEPLVTQ VTTLVNTNSK GPSNKKRGRS KKAHVLAASV EQATENFLEK GDKIAKESQF LKEELVAAVE DVRKQGDLMK AAAGEFADDP CSSVKRGNMV RAARALLSAV TRLLILADMA DVYKLLVQLK VVEDGILKLR NAGNEQDLGI QYKALKPEVD KLNIMAAKRQ QELKDVGHRD QMAAARGILQ KNVPILYTAS QACLQHPDVA AYKANRDLIY KQLQQAVTGI SNAAQATASD DASQHQGGGG GELAYALNNF DKQIIVDPLS FSEERFRPSL EERLESIISG AALMADSSCT RDDRRERIVA ECNAVRQALQ DLLSEYMGNA GRKERSDALN SAIDKMTKKT RDLRRQLRKA VMDHVSDSFL ETNVPLLVLI EAAKNGNEKE VKEYAQVFRE HANKLIEVAN LACSISNNEE GVKLVRMSAS QLEALCPQVI NAALALAAKP QSKLAQENMD LFKEQWEKQV RVLTDAVDDI TSIDDFLAVS ENHILEDVNK CVIALQEKDV DGLDRTAGAI RGRAARVIHV VTSEMDNYEP GVYTEKVLEA TKLLSNTVMP RFTEQVEAAV EALSSDPAQP MDENEFIDAS RLVYDGIRDI RKAVLMIRTP EELDDSDFET EDFDVRSRTS VQTEDDQLIA GQSARAIMAQ LPQEQKAKIA EQVASFQEEK SKLDAEVSKW DDSGNDIIVL AKQMCMIMME MTDFTRGKGP LKNTSDVISA AKKIAEAGSR MDKLGRTIAD HCPDSACKQD LLAYLQRIAL YCHQLNICSK VKAEVQNLGG ELVVSGVDSA MSLIQAAKNL MNAVVQTVKA SYVASTKYQK SQGMASLNLP AVSWKMKAPE KKPLVKREKQ DETQTKIKRA SQKKHVNPVQ ALSEFKAMDS I |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.25 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 294.15 K / Instrument: LEICA EM GP |
- Electron microscopy
Electron microscopy
| Microscope | JEOL CRYO ARM 300 |
|---|---|
| Specialist optics | Phase plate: OTHER / Energy filter - Name: In-column Omega Filter / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average exposure time: 0.1 sec. / Average electron dose: 48.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 60000 |
| Sample stage | Specimen holder model: JEOL CRYOSPECPORTER / Cooling holder cryogen: NITROGEN |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT |
|---|
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)